Building RAG-based LLM Applications for Production
Check out our updated RAG blog
For the most up-to-date content on how to run the best RAG pipelines with Ray, read our updated blog.
[ GitHub | Notebook | Anyscale Endpoints | Ray Docs] · 55 min read
Note: Check out the new evaluation reports and cost analysis with mixtral-8x7b-instruct-v0.1 and our data flywheel workflow to continuously improve our RAG applications.
In this guide, we will learn how to:
💻 Develop a retrieval augmented generation (RAG) based LLM application from scratch.
🚀 Scale the major workloads (load, chunk, embed, index, serve, etc.) across multiple workers with different compute resources.
✅ Evaluate different configurations of our application to optimize for both per-component (ex.
retrieval_score) and overall performance (quality_score).🔀 Implement a hybrid agent routing approach b/w OSS and closed LLMs to create the most performant and cost effective application.
📦 Serve the application in a highly scalable and available manner.
💡 Learn how methods like fine-tuning, prompt engineering, lexical search, reranking, data flywheel, etc. impact our application's performance.
LinkOverview
Large language models (LLMs) have undoubtedly changed the way we interact with information. However, they come with their fair share of limitations as to what we can ask of them. Base LLMs (ex. Llama-2-70b, gpt-4, etc.) are only aware of the information that they've been trained on and will fall short when we require them to know information beyond that. Retrieval augmented generation (RAG) based LLM applications address this exact issue and extend the utility of LLMs to our specific data sources.
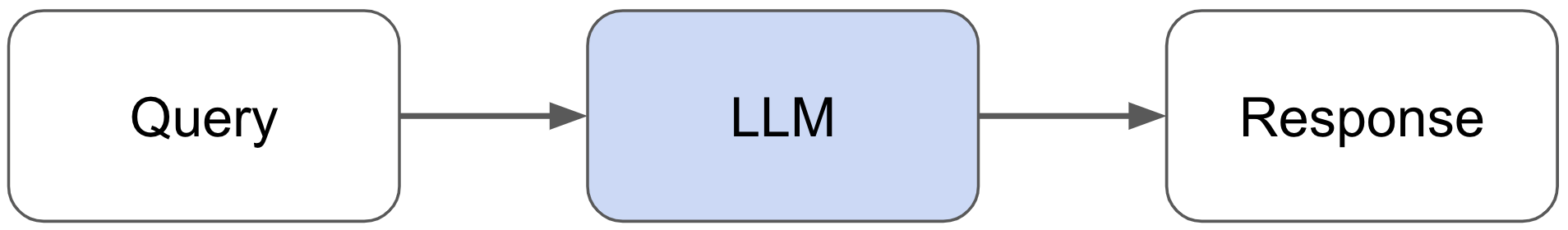
In this guide, we're going to build a RAG-based LLM application where we will incorporate external data sources to augment our LLM’s capabilities. Specifically, we will be building an assistant that can answer questions about Ray — a Python framework for productionizing and scaling ML workloads. The goal here is to make it easier for developers to adopt Ray, but also, as we'll see in this guide, to help improve our Ray documentation itself and provide a foundation for other LLM applications. We’ll also share challenges we faced along the way and how we overcame them.
Note: We have generalized this entire guide so that it can easily be extended to build RAG-based LLM applications on top of your own data.
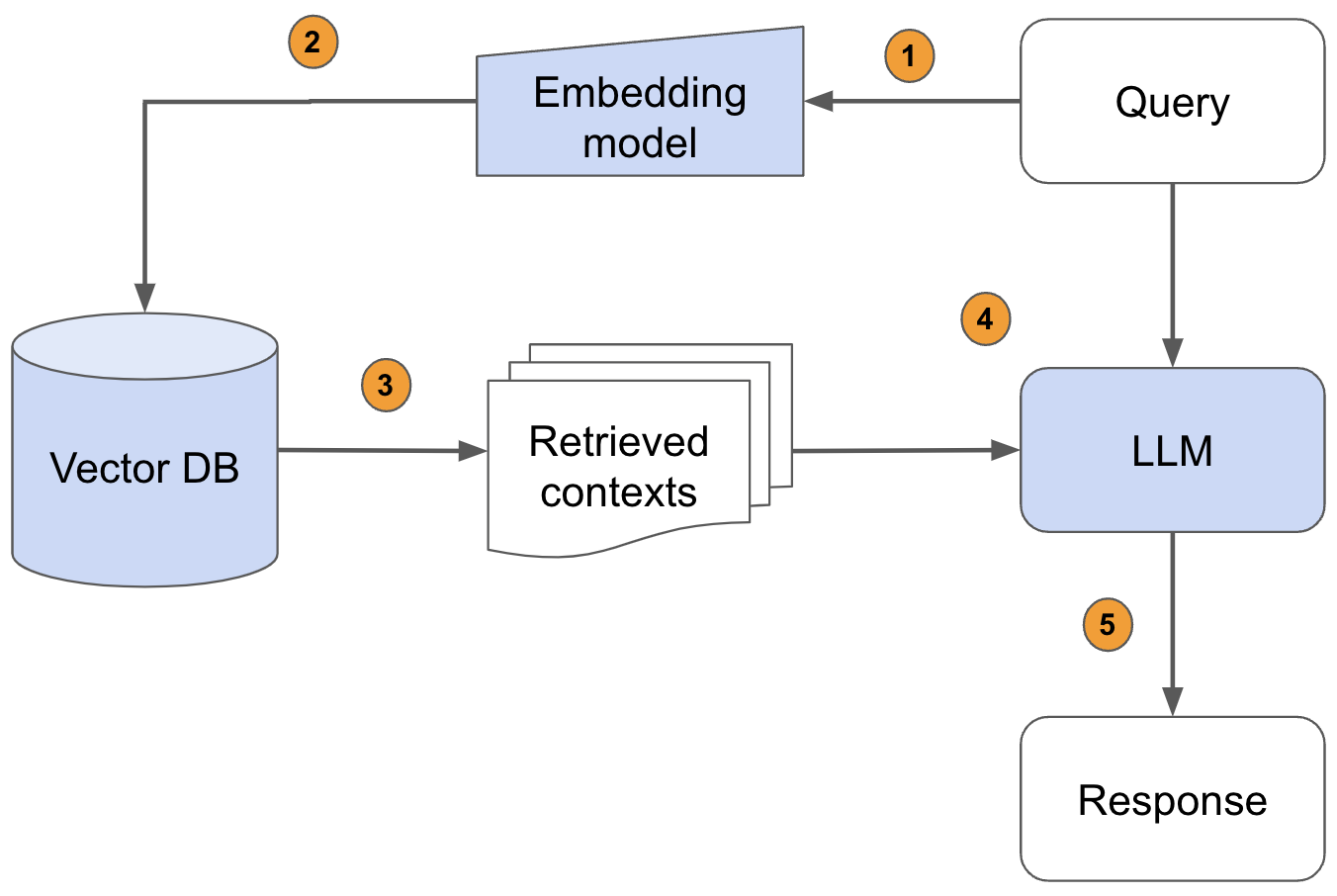
Pass the query to the embedding model to semantically represent it as an embedded query vector.Pass the embedded query vector to our vector DB.Retrieve the top-k relevant contexts – measured by distance between the query embedding and all the embedded chunks in our knowledge base.Pass the query text and retrieved context text to our LLM.The LLM will generate a response using the provided content.
Besides just building our LLM application, we’re also going to be focused on scaling and serving it in production. Unlike traditional machine learning, or even supervised deep learning, scale is a bottleneck for LLM applications from the very beginning. Large datasets, models, compute intensive workloads, serving requirements, etc. We’ll develop our application to be able to handle any scale as the world around us continues to grow.
We’re also going to be focused on evaluation and performance. Our application involves many moving pieces: embedding models, chunking logic, the LLM itself, etc. and so it's important that we experiment with different configurations to optimize for the best quality responses. However, it's non-trivial to evaluate and quantitatively compare different configurations for a generative task. We’re going to break down evaluation of individual parts of our application (retrieval given query, generation given source), also assess the overall performance (end-to-end generation) and share findings towards an optimized configuration.
Note: We'll be experimenting with different LLMs (OpenAI, Llama, etc.) in this guide. You will need OpenAI credentials to access ChatGPT models and Anyscale Endpoints (public and private endpoints available) to serve + fine-tune OSS LLMs.
LinkVector DB creation
Before we can start building our RAG application, we need to first create our vector DB that will contain our processed data sources.
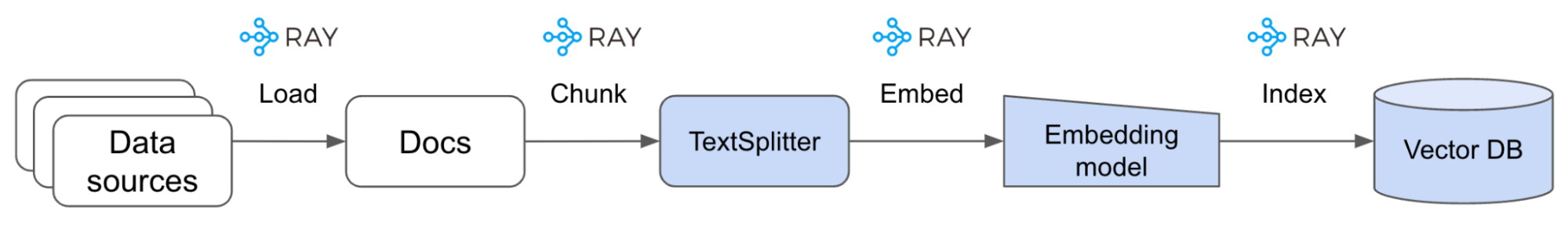
LinkLoad data
We’re going to start by loading the Ray documentation from the website to a local directory:
1export EFS_DIR=/desired/output/directory
2wget -e robots=off --recursive --no-clobber --page-requisites \
3 --html-extension --convert-links --restrict-file-names=windows \
4 --domains docs.ray.io --no-parent --accept=html \
5 -P $EFS_DIR https://docs.ray.io/en/master/We’re going to then load our docs contents into a Ray Dataset so that we can perform operations at scale on them (ex. embed, index, etc.). With large data sources, models and application serving needs, scale is a day-1 priority for LLM applications. We want to build our applications in such a way that they can scale as our needs grow without us having to change our code later.
1# Ray dataset
2DOCS_DIR = Path(EFS_DIR, "docs.ray.io/en/master/")
3ds = ray.data.from_items([{"path": path} for path in DOCS_DIR.rglob("*.html")
4if not path.is_dir()])
5print(f"{ds.count()} documents")LinkSections
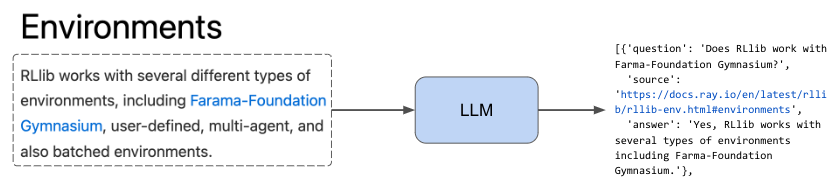
Now that we have a dataset of all the paths to the html files, we're going to develop some functions that can appropriately extract the content from these files. We want to do this in a generalized manner so that we can perform this extraction across all of our docs pages (and so you can use it for your own data sources). Our process is to first identify the sections in our html page and then extract the text in between them. We save all of this into a list of dictionaries that map the text within a section to a specific url with a section anchor id.
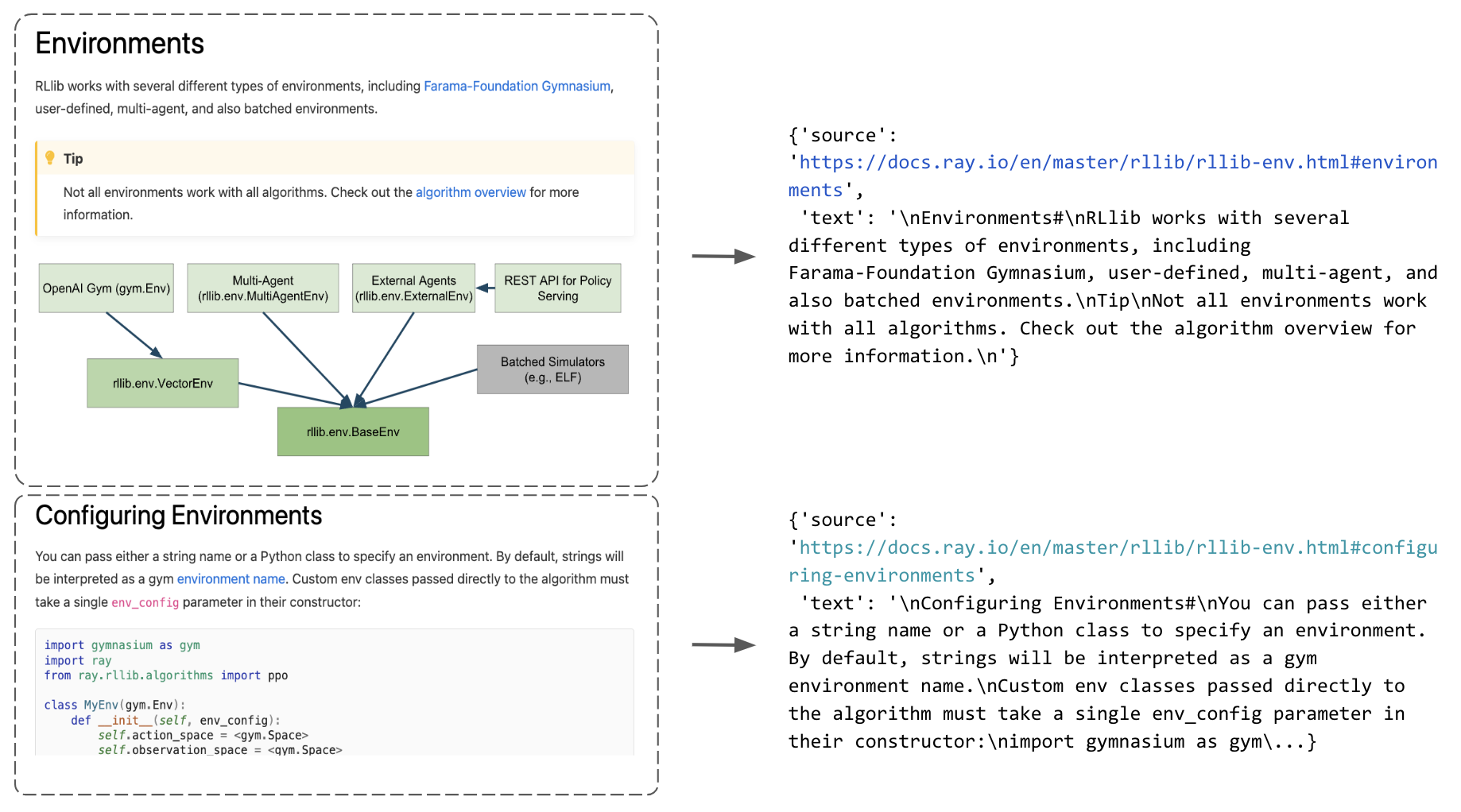
1sample_html_fp = Path(EFS_DIR, "docs.ray.io/en/master/rllib/rllib-env.html")
2extract_sections({"path": sample_html_fp})[0]
3{'source': 'https://docs.ray.io/en/master/rllib/rllib-env.html#environments', 'text': '\nEnvironments#\nRLlib works with several different types of environments, including Farama-Foundation Gymnasium, user-defined, multi-agent, and also batched environments.\nTip\nNot all environments work with all algorithms. Check out the algorithm overview for more information.\n'}
We can apply this extraction process (extract_section) in parallel to all the file paths in our dataset with just one line using Ray Data’s flat_map.
1# Extract sections
2sections_ds = ds.flat_map(extract_sections)
3sections = sections_ds.take_all()
4print (len(sections))
5LinkChunk data
We now have a list of sections (with text and source of each section) but we shouldn't directly use this as context to our RAG application just yet. The text lengths of each section are all varied and many are quite large chunks.
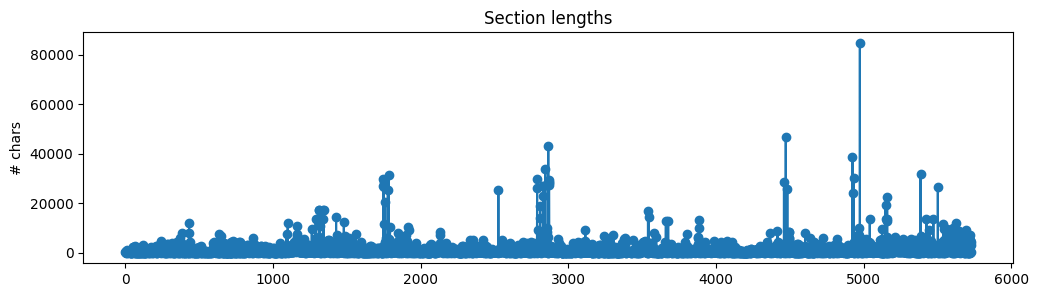
If we were to use these large sections, then we'd be inserting a lot of noisy/unwanted context and because all LLMs have a maximum context length, we wouldn't be able to fit too much other relevant context. So instead, we're going to split the text within each section into smaller chunks. Intuitively, smaller chunks will encapsulate single/few concepts and will be less noisy compared to larger chunks. We're going to choose some typical text splitting values (ex. chunk_size=300) to create our chunks for now but we'll be experimenting with a wider range of values later.
1from langchain.document_loaders import ReadTheDocsLoader
2from langchain.text_splitter import RecursiveCharacterTextSplitter
3
4# Text splitter
5chunk_size = 300
6chunk_overlap = 50
7text_splitter = RecursiveCharacterTextSplitter(
8 separators=["\n\n", "\n", " ", ""],
9 chunk_size=chunk_size,
10 chunk_overlap=chunk_overlap,
11 length_function=len,
12)
13
14# Chunk a sample section
15sample_section = sections_ds.take(1)[0]
16chunks = text_splitter.create_documents(
17 texts=[sample_section["text"]],
18 metadatas=[{"source": sample_section["source"]}])
19print (chunks[0])
20page_content='ray.tune.TuneConfig.search_alg#\nTuneConfig.search_alg: Optional[Union[ray.tune.search.searcher.Searcher, ray.tune.search.search_algorithm.SearchAlgorithm]] = None#' metadata={'source': 'https://docs.ray.io/en/master/tune/api/doc/ray.tune.TuneConfig.search_alg.html#ray-tune-tuneconfig-search-alg'}
While chunking our dataset is relatively fast, let’s wrap the chunking logic into a function so that we can apply the workload at scale so that chunking remains just as fast as our data sources grow:
1def chunk_section(section, chunk_size, chunk_overlap):
2 text_splitter = RecursiveCharacterTextSplitter(
3 separators=["\n\n", "\n", " ", ""],
4 chunk_size=chunk_size,
5 chunk_overlap=chunk_overlap,
6 length_function=len)
7 chunks = text_splitter.create_documents(
8 texts=[sample_section["text"]],
9 metadatas=[{"source": sample_section["source"]}])
10 return [{"text": chunk.page_content, "source": chunk.metadata["source"]} for chunk in chunks]
11
12# Scale chunking
13chunks_ds = sections_ds.flat_map(partial(
14 chunk_section,
15 chunk_size=chunk_size,
16 chunk_overlap=chunk_overlap))
17print(f"{chunks_ds.count()} chunks")
18chunks_ds.show(1)
195727 chunks
{'text': 'ray.tune.TuneConfig.search_alg#\nTuneConfig.search_alg: Optional[Union[ray.tune.search.searcher.Searcher, ray.tune.search.search_algorithm.SearchAlgorithm]] = None#', 'source': 'https://docs.ray.io/en/master/tune/api/doc/ray.tune.TuneConfig.search_alg.html#ray-tune-tuneconfig-search-alg'}
LinkEmbed data
Now that we've created small chunks from our sections, we need a way to identify the most relevant ones for a given query. A very effective and quick method is to embed our data using a pretrained model and use the same model to embed the query. We can then compute the distance between all of the chunk embeddings and our query embedding to determine the top-k chunks. There are many different pretrained models to choose from to embed our data but the most popular ones can be discovered through HuggingFace's Massive Text Embedding Benchmark (MTEB) leaderboard. These models were pretrained on very large text corpus through tasks such as next/masked token prediction which allowed them to learn to represent sub-tokens in N dimensions and capture semantic relationships. We can leverage this to represent our data and identify the most relevant contexts to use to answer a given query. We're using Langchain's Embedding wrappers (HuggingFaceEmbeddings and OpenAIEmbeddings) to easily load the models and embed our document chunks.
Note: embeddings aren't the only way to determine the more relevant chunks. We could also use an LLM to decide! However, because LLMs are much larger than these embedding models and have maximum context lengths, it's better to use embeddings to retrieve the top k chunks. And then we could use LLMs on the fewer k chunks to determine the <k chunks to use as the context to answer our query. We could also use reranking (ex. Cohere Rerank) to further identify the most relevant chunks to use. We could also combine embeddings with traditional information retrieval methods such as keyword matching, which could be useful for matching for unique tokens that may potentially be lost when embedding sub-tokens.
1from langchain.embeddings import OpenAIEmbeddings
2from langchain.embeddings.huggingface import HuggingFaceEmbeddings
3import numpy as np
4from ray.data import ActorPoolStrategy
5
6class EmbedChunks:
7 def __init__(self, model_name):
8 if model_name == "text-embedding-ada-002":
9 self.embedding_model = OpenAIEmbeddings(
10 model=model_name,
11 openai_api_base=os.environ["OPENAI_API_BASE"],
12 openai_api_key=os.environ["OPENAI_API_KEY"])
13 else:
14 self.embedding_model = HuggingFaceEmbeddings(
15 model_name=model_name,
16 model_kwargs={"device": "cuda"},
17 encode_kwargs={"device": "cuda", "batch_size": 100})
18
19 def __call__(self, batch):
20 embeddings = self.embedding_model.embed_documents(batch["text"])
21 return {"text": batch["text"], "source": batch["source"], "embeddings":
22embeddings}
23Here we're able to embed our chunks at scale by using map_batches. All we had to do was define the batch_size and the compute (we're using two workers, each with 1 GPU).
1# Embed chunks
2embedding_model_name = "thenlper/gte-base"
3embedded_chunks = chunks_ds.map_batches(
4 EmbedChunks,
5 fn_constructor_kwargs={"model_name": embedding_model_name},
6 batch_size=100,
7 num_gpus=1,
8 compute=ActorPoolStrategy(size=2))
9# Sample (text, source, embedding) triplet
[{'text': 'External library integrations for Ray Tune#',
'source': 'https://docs.ray.io/en/master/tune/api/integration.html#external-library-integrations-for-ray-tune',
'embeddings': [
0.012108353897929192,
0.009078810922801495,
0.030281754210591316,
-0.0029687234200537205,
…]
}
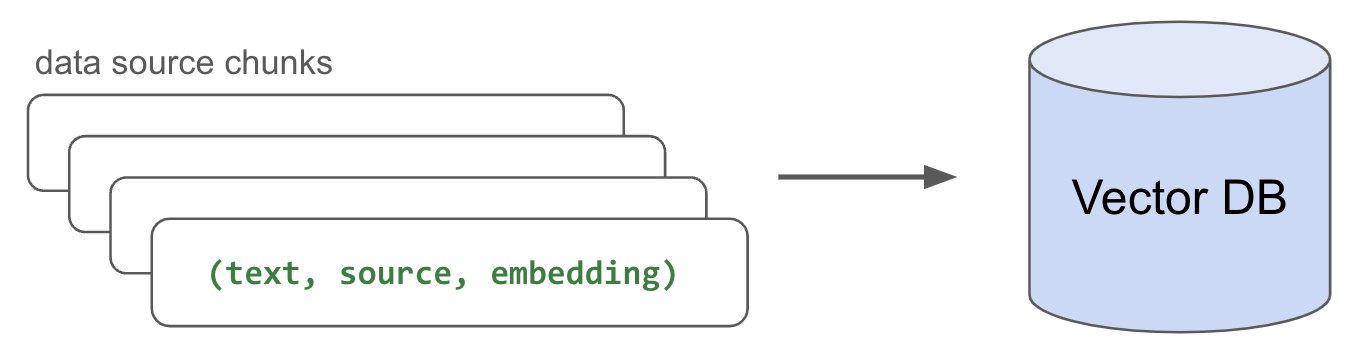
LinkIndex data
Now that we have our embedded chunks, we need to index (store) them somewhere so that we can retrieve them quickly for inference. While there are many popular vector database options, we're going to use Postgres with pgvector for its simplicity and performance. We'll create a table (document) and write the (text, source, embedding) triplets for each embedded chunk we have.
1class StoreResults:
2 def __call__(self, batch):
3 with psycopg.connect(os.environ["DB_CONNECTION_STRING"]) as conn:
4 register_vector(conn)
5 with conn.cursor() as cur:
6 for text, source, embedding in zip
7 (batch["text"], batch["source"], batch["embeddings"]):
8 cur.execute("INSERT INTO document (text, source, embedding)
9 VALUES (%s, %s, %s)", (text, source, embedding,),)
10 return {}
11And once again, we can use Ray Data’s map_batches to perform this indexing in parallel:
1# Index data
2embedded_chunks.map_batches(
3 StoreResults,
4 batch_size=128,
5 num_cpus=1,
6 compute=ActorPoolStrategy(size=28),
7).count()
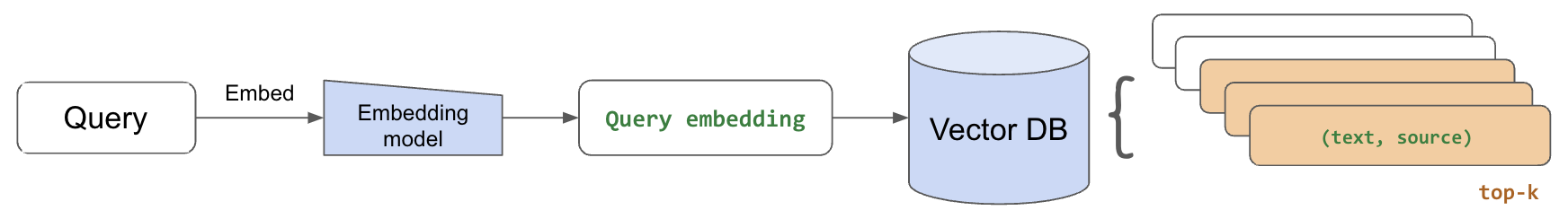
8LinkQuery Retrieval
With our embedded chunks indexed in our vector database, we're ready to perform retrieval for a given query. We'll start by using the same embedding model we used to embed our text chunks to now embed the incoming query.
1# Embed query
2embedding_model = HuggingFaceEmbeddings(model_name=embedding_model_name)
3query = "What is the default batch size for map_batches?"
4
5embedding = np.array(embedding_model.embed_query(query))
6len(embedding)
7768
Then, we'll retrieve the top-k most relevant chunks by extracting the closest embedded chunks to our embedded query. We use cosine distance (<=>) but there are many options to choose from. Once we retrieve the top num_chunks, we can collect the text for each chunk and use it as context to generate a response.
1# Get context
2num_chunks = 5
3with psycopg.connect(os.environ["DB_CONNECTION_STRING"]) as conn:
4 register_vector(conn)
5 with conn.cursor() as cur:
6 cur.execute("SELECT * FROM document ORDER BY embedding <-> %s LIMIT %s", (embedding, num_chunks))
7 rows = cur.fetchall()
8 context = [{"text": row[1]} for row in rows]
9 sources = [row[2] for row in rows]
10https://docs.ray.io/en/master/data/api/doc/ray.data.Dataset.map_batches.html#ray-data-dataset-map-batches
entire blocks as batches (blocks may contain different numbers of rows).
The actual size of the batch provided to fn may be smaller than
batch_size if batch_size doesn’t evenly divide the block(s) sent
to a given map task. Default batch_size is 4096 with “default”.
https://docs.ray.io/en/master/data/transforming-data.html#configuring-batch-size
The default batch size depends on your resource type. If you’re using CPUs,
the default batch size is 4096. If you’re using GPUs, you must specify an explicit batch size.
(cont…)
And we can combine all of this into one convenient function:
1def semantic_search(query, embedding_model, k):
2 embedding = np.array(embedding_model.embed_query(query))
3 with psycopg.connect(os.environ["DB_CONNECTION_STRING"]) as conn:
4 register_vector(conn)
5 with conn.cursor() as cur:
6 cur.execute("SELECT * FROM document ORDER BY embedding <=> %s LIMIT %s", (embedding, k),)
7 rows = cur.fetchall()
8 semantic_context = [{"id": row[0], "text": row[1], "source": row[2]} for row in rows]
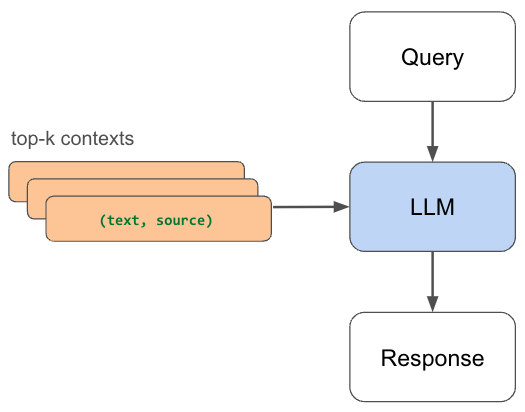
9 return semantic_contextLinkResponse Generation
We can now use the context to generate a response from our LLM. Without this relevant context that we retrieved, the LLM may not have been able to accurately answer our question. And as our data grows, we can just as easily embed and index any new data and be able to retrieve it to answer questions.
1from rag.generate import prepare_response
2from rag.utils import get_client
3
4def generate_response(
5 llm, temperature=0.0, stream=True,
6 system_content="", assistant_content="", user_content="",
7 max_retries=1, retry_interval=60):
8 """Generate response from an LLM."""
9 retry_count = 0
10 client = get_client(llm=llm)
11 messages = [{"role": role, "content": content} for role, content in [
12 ("system", system_content),
13 ("assistant", assistant_content),
14 ("user", user_content)] if content]
15 while retry_count <= max_retries:
16 try:
17 chat_completion = client.chat.completions.create(
18 model=llm,
19 temperature=temperature,
20 stream=stream,
21 messages=messages,
22 )
23 return prepare_response(chat_completion, stream=stream)
24
25 except Exception as e:
26 print(f"Exception: {e}")
27 time.sleep(retry_interval) # default is per-minute rate limits
28 retry_count += 1
29 return ""Note: We’re using a temperature of 0.0 to enable reproducible experiments but you should adjust this based on your use case. For use cases that need to always be factually grounded, we recommend very low temperature values while more creative tasks can benefit from higher temperatures.
1# Generate response
2query = "What is the default batch size for map_batches?"
3response = generate_response(
4 llm="meta-llama/Llama-2-70b-chat-hf",
5 temperature=0.0,
6 stream=True,
7 system_content="Answer the query using the context provided. Be succinct.",
8 user_content=f"query: {query}, context: {context}")
9The default batch size for map_batches is 4096.
LinkAgent
Let's combine the context retrieval and response generation together into a convenient query agent that we can use to easily generate our responses. This will take care of setting up our agent (embedding and LLM model), as well as the context retrieval, and pass it to our LLM for response generation.
1class QueryAgent:
2 def __init__(self, embedding_model_name="thenlper/gte-base",
3 llm="meta-llama/Llama-2-70b-chat-hf", temperature=0.0,
4 max_context_length=4096, system_content="", assistant_content=""):
5
6 # Embedding model
7 self.embedding_model = get_embedding_model(
8 embedding_model_name=embedding_model_name,
9 model_kwargs={"device": "cuda"},
10 encode_kwargs={"device": "cuda", "batch_size": 100})
11
12 # Context length (restrict input length to 50% of total length)
13 max_context_length = int(0.5*max_context_length)
14
15 # LLM
16 self.llm = llm
17 self.temperature = temperature
18 self.context_length = max_context_length - get_num_tokens(system_content + assistant_content)
19 self.system_content = system_content
20 self.assistant_content = assistant_content
21
22 def __call__(self, query, num_chunks=5, stream=True):
23 # Get sources and context
24 context_results = semantic_search(
25 query=query,
26 embedding_model=self.embedding_model,
27 k=num_chunks)
28
29 # Generate response
30 context = [item["text"] for item in context_results]
31 sources = [item["source"] for item in context_results]
32 user_content = f"query: {query}, context: {context}"
33
34 answer = generate_response(
35 llm=self.llm,
36 temperature=self.temperature,
37 stream=stream,
38 system_content=self.system_content,
39 assistant_content=self.assistant_content,
40 user_content=trim(user_content, self.context_length))
41
42 # Result
43 result = {
44 "question": query,
45 "sources": sources,
46 "answer": answer,
47 "llm": self.llm,
48 }
49 return resultWith this, we can use our RAG application in just a few lines:
1llm = "meta-llama/Llama-2-7b-chat-hf"
2agent = QueryAgent(
3 embedding_model_name="thenlper/gte-base",
4 llm=llm,
5 max_context_length=MAX_CONTEXT_LENGTHS[llm],
6 system_content="Answer the query using the context provided. Be succinct.")
7result = agent(query="What is the default batch size for map_batches?")
8print("\n\n", json.dumps(result, indent=2))The default batch size for `map_batches` is 4096
{
"question": "What is the default batch size for map_batches?",
"sources": [
"ray.data.Dataset.map_batches — Ray 2.7.1",
"Transforming Data — Ray 2.7.1",
"Ray Data Internals — Ray 2.7.1",
"Dynamic Request Batching — Ray 2.7.1",
"Image Classification Batch Inference with PyTorch — Ray 2.7.1"
],
"answer": "The default batch size for `map_batches` is 4096",
"llm": "meta-llama/Llama-2-7b-chat-hf"
}
LinkEvaluation
So far, we've chosen typical/arbitrary values for the various parts of our RAG application. But if we were to change something, such as our chunking logic, embedding model, LLM, etc. how can we know that we have a better configuration than before? A generative task like this is very difficult to quantitatively assess and so we need to develop reliable ways to do so.
Because we have many moving parts in our application, we need to perform both unit/component and end-to-end evaluation. Component-wise evaluation can involve evaluating our retrieval in isolation (is the best source in our set of retrieved chunks) and evaluating our LLMs response (given the best source, is the LLM able to produce a quality answer). And for end-to-end evaluation, we can assess the quality of the entire system (given the data sources, what is the quality of the response).
We'll be asking our evaluator LLM to score the quality of the response between 1-5 using the context, however, we could also have it produce scores for other dimensions such as hallucination (is the generated answer using information only from the provided context), toxicity, etc.
Note: We could have constrained the score to be binary (0/1), which might be more interpretable (ex. the response was either correct or incorrect). However, we introduced a higher variance in our scores to develop a deeper, fine-grained, understanding of how LLMs score responses (ex. LLM bias towards responses).
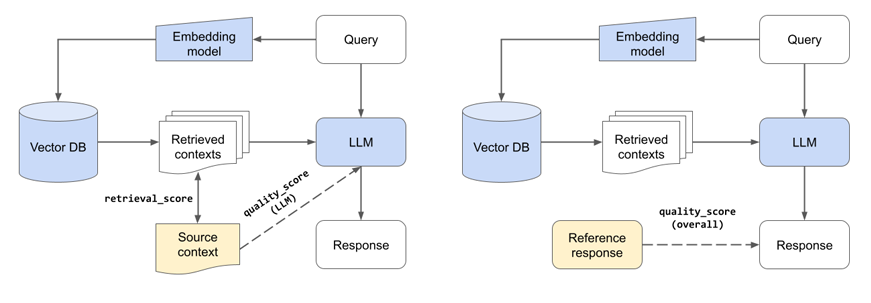
Component evaluations (left) of retrieval system and LLM. Overall evaluation (right).
LinkEvaluator
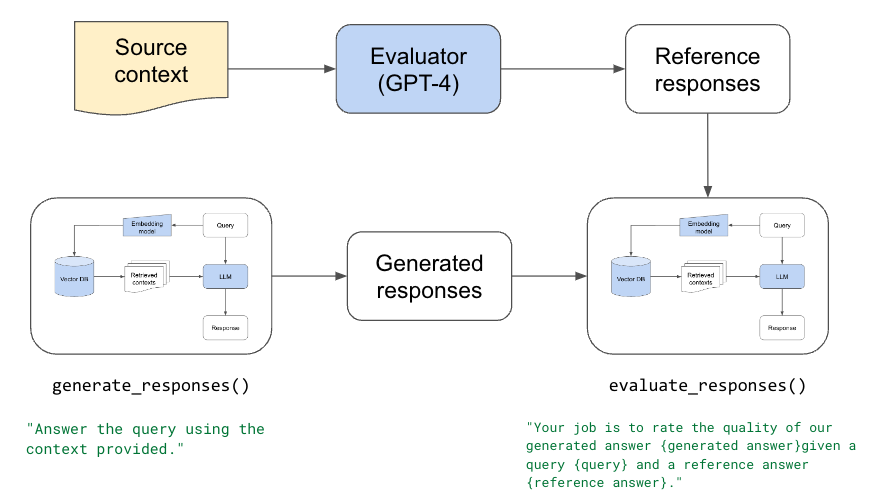
We're going to start by determining our evaluator. Given a response to a query and relevant context, our evaluator should be a trusted way to score/assess the quality of the response. But before we can determine our evaluator, we need a dataset of questions and the source where the answer comes from. We can use this dataset to ask our different evaluators to provide an answer and then rate their answer (ex. score between 1-5). We can then inspect this dataset to determine if our evaluator is unbiased and has sound reasoning for the scores that are assigned.
Note: We’re evaluating the ability of our LLM to generate a response given the relevant context. This is a component-level evaluation (quality_score (LLM)) because we aren’t using retrieval to fetch the relevant context.
We'll start by manually creating our dataset (keep reading if you can’t manually create a dataset). We have a list of user queries and the ideal source to answer the query datasets/eval-dataset-v1.jsonl. We will use our LLM app above to generate reference answers for each query/source pair using gpt-4.
1with open(Path(ROOT_DIR, "datasets/eval-dataset-v1.jsonl"), "r") as f:
2 data = [json.loads(item) for item in list(f)]
3[{'question': 'I’m struggling a bit with Ray Data type conversions when I do map_batches. Any advice?',
'source': 'https://docs.ray.io/en/master/data/transforming-data.html'},
…
{'question': 'Is Ray integrated with DeepSpeed?',
'source': 'https://docs.ray.io/en/master/ray-air/examples/gptj_deepspeed_fine_tuning.html#fine-tuning-the-model-with-ray-air-a-name-train-a'}]
Each data point has a question and the labeled source that has the precise context with the answer to the question:
1# Sample
2uri = "https://docs.ray.io/en/master/data/transforming-data.html#configuring-batch-format"
3fetch_text(uri=uri)
4'\nConfiguring batch format#\nRay Data represents batches as dicts of NumPy ndarrays or pandas DataFrames. …'
We can extract the text from this context and pass it to our LLM to generate a response to the question. We’re also going to ask it to score the quality of its response for the query. To do this, we’ve defined a QueryAgentWithContext that inherits from QueryAgent, with the change that we’re providing the context and it doesn’t need to retrieve it.
1class QueryAgentWithContext(QueryAgent):
2 def __call__(self, query, context):
3 user_content = f"query: {query}, context: {context}"
4 response = generate_response(
5 llm=self.llm,
6 temperature=self.temperature,
7 stream=True,
8 system_content=self.system_content,
9 assistant_content=self.assistant_content,
10 user_content=user_content[: self.context_length])
11 return response
12We can now create a dataset with question, source, answer, score and reasoning. We can inspect this to determine if our evaluator is of high quality.
question:"I’m struggling a bit with Ray Data type conversions when I do map_batches.
Any advice?"
source:"https://docs.ray.io/en/master/data/transforming-data.html#configuring-batch-format"
answer:"You can configure the batch type in Ray Data by specifying the 'batch_format' in the 'map_batches()' function. If you're working with NumPy ndarrays, your function should return a dict of ndarrays. If you're working with pandas DataFrames, your function should return a DataFrame. Make sure your function is returning the correct type based on your specified 'batch_format'."
score:5
reasoning:"The context provides clear instructions on how to configure the batch type in Ray Data and how to use the 'map_batches()' function. It also provides examples for both NumPy and pandas, which directly answers the query."
We found that gpt-4 was a high quality evaluator based on the scores and reasonings it provided. We performed the same evaluation with other LLMs (ex. Llama-2-70b) and we found that they lacked the appropriate reasoning and were very generous with responses from themselves.
1EVALUATOR = "gpt-4"Note: A more thorough evaluation would also test for the following by asking the evaluator to compare responses from different LLMs across the following:
position (which responses we show first)
verbosity (longer responses are favored)
nepotism (ex. GPT4 prefers GPT 3.5, etc.)
LinkCold Start
We may not always have a prepared dataset of questions and the best source to answer that question readily available. To address this cold start problem, we could use an LLM to look at our text chunks and generate questions that the specific chunk would answer. This provides us with quality questions and the exact source the answer is in. However, this dataset generation method could be a bit noisy. The generated questions may not always have high alignment to what our users may ask. And the specific chunk we say is the best source may also have that exact information in other chunks. Nonetheless, this is a great way to start our development process while we collect + manually label a high quality dataset.
1# Prompt
2num_questions = 3
3system_content = f"""
4Create {num_questions} questions using only the context provided.
5End each question with a '?' character and then in a newline write the answer to that question using only the context provided.
6Separate each question/answer pair by a newline.
7"""
8
9# Generate questions
10synthetic_data = []
11for chunk in chunks[:1]: # small samples
12 response = generate_response(
13 llm="gpt-4",
14 temperature=0.0,
15 system_content=system_content,
16 user_content=f"context: {chunk.page_content}")
17 entries = response.split("\n\n")
18 for entry in entries:
19 question, answer = entry.split("\n")
20 synthetic_data.append({"question": question, "source": chunk.metadata["source"], "answer": answer})
21synthetic_data[:3]
22[{'question': 'What can you use to monitor and debug your Ray applications and clusters?',
'source': 'https://docs.ray.io/en/master/ray-observability/reference/index.html#reference',
'answer': 'You can use the API and CLI documented in the references to monitor and debug your Ray applications and clusters.'},
{'question': 'What are the guides included in the references?',
'source': 'https://docs.ray.io/en/master/ray-observability/reference/index.html#reference',
'answer': 'The guides included in the references are State API, State CLI, and System Metrics.'},
{'question': 'What are the two types of interfaces mentioned for monitoring and debugging Ray applications and clusters?',
'source': 'https://docs.ray.io/en/master/ray-observability/reference/index.html#reference',
'answer': 'The two types of interfaces mentioned for monitoring and debugging Ray applications and clusters are API and CLI.'}]
LinkLLM Experiments
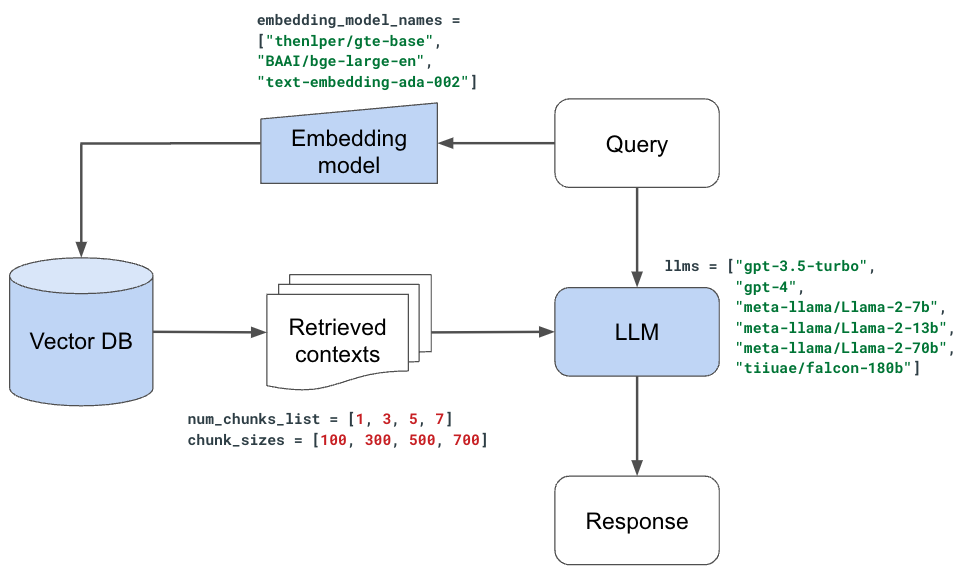
With our evaluator set, we're ready to start experimenting with the various components in our LLM application. While we could perform this as a large hyperparameter tuning experiment, where we can search across promising combinations of values/decisions, we're going to evaluate one decision at a time and set the best value for the next experiment.
Note: this approach is slightly imperfect because many of our decisions are not independent (ex. chunk_size and num_chunks should ideally be evaluated across many combinations of values).
LinkUtilities
Before we start our experiments, we’re going to define a few more utility functions. Our evaluation workflow will use our evaluator to assess the end-to-end quality (quality_score (overall)) of our application since the response depends on the retrieved context and the LLM. But we’ll also include a retrieval_score to measure the quality of our retrieval process (chunking + embedding). Our logic for determining the retrieval_score registers a success if the best source is anywhere in our retrieved num_chunks sources. We don't account for order, exact page section, etc. but we could add those constraints to have a more conservative retrieval score.
1def get_retrieval_score(references, generated):
2 matches = np.zeros(len(references))
3 for i in range(len(references)):
4 reference_source = references[i]["source"].split("#")[0]
5 if not reference_source:
6 matches[i] = 1
7 continue
8 for source in generated[i]["sources"]:
9 # sections don't have to perfectly match
10 if reference_source == source.split("#")[0]:
11 matches[i] = 1
12 continue
13 retrieval_score = np.mean(matches)
14 return retrieval_score
15Regardless of what configuration(s) we want to evaluate, we’ll need to first generate responses using that configuration and then evaluate those responses using our evaluator:
1# Generate responses
2generation_system_content = "Answer the query using the context provided. Be succinct."
3generate_responses(
4 experiment_name=experiment_name,
5 chunk_size=chunk_size,
6 chunk_overlap=chunk_overlap,
7 num_chunks=num_chunks,
8 embedding_model_name=embedding_model_name,
9 use_lexical_search=use_lexical_search,
10 lexical_search_k=lexical_search_k,
11 use_reranking=use_reranking,
12 rerank_threshold=rerank_threshold,
13 rerank_k=rerank_k,
14 llm=llm,
15 temperature=0.0,
16 max_context_length=MAX_CONTEXT_LENGTHS[llm],
17 system_content=generation_system_content,
18 assistant_content="",
19 docs_dir=docs_dir,
20 experiments_dir=experiments_dir,
21 references_fp=references_fp,
22 num_samples=num_samples,
23 sql_dump_fp=sql_dump_fp)1# Evaluate responses
2evaluation_system_content = """
3 Your job is to rate the quality of our generated answer {generated_answer}
4 given a query {query} and a reference answer {reference_answer}.
5 Your score has to be between 1 and 5.
6 You must return your response in a line with only the score.
7 Do not return answers in any other format.
8 On a separate line provide your reasoning for the score as well.
9 """
10evaluate_responses(
11 experiment_name=experiment_name,
12 evaluator=evaluator,
13 temperature=0.0,
14 max_context_length=MAX_CONTEXT_LENGTHS[evaluator],
15 system_content=evaluation_system_content,
16 assistant_content="",
17 experiments_dir=experiments_dir,
18 references_fp=references_fp,
19 responses_fp=str(Path(experiments_dir, "responses", f"{experiment_name}.json")),
20 num_samples=num_samples)
21We combine both of these steps into a convenient run_experiment function:
1# Run experiment
2run_experiment(
3 experiment_name=experiment_name,
4 chunk_size=CHUNK_SIZE,
5 chunk_overlap=CHUNK_OVERLAP,
6 num_chunks=NUM_CHUNKS,
7 embedding_model_name=EMBEDDING_MODEL_NAME,
8 llm=LLM,
9 evaluator=EVALUATOR,
10 docs_dir=DOCS_DIR,
11 experiments_dir=EXPERIMENTS_DIR,
12 references_fp=REFERENCES_FILE_PATH,
13 num_samples=NUM_SAMPLES)Note: We won’t crowd this blog post with all the code to run each experiment but you can find all of it on our GitHub repository.
LinkContext
We’re now ready to start our experiments! We're going to first test if the additional context we provide is helpful at all. This is to validate that the RAG system is indeed worth the effort. We can do this by setting num_chunks=0 (no context) and comparing that to num_chunks=5.
1# Without context
2num_chunks = 0
3experiment_name = f"without-context"
4run_experiment()
5
6# With context
7num_chunks = 5
8experiment_name = f"with-context"
9run_experiment()
10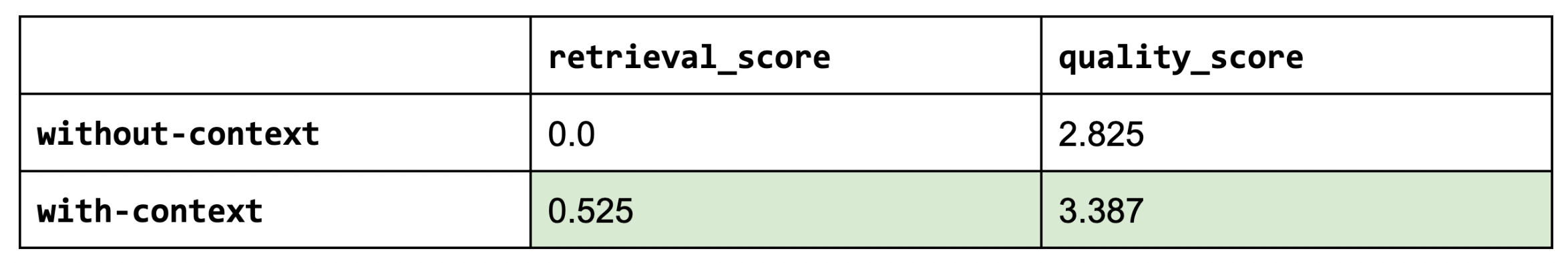
Sanity check: the retrieval score for without-context is zero since we’re using any context.
As we can see, using context (RAG) does indeed help in the quality of our answers (and by a meaningful margin).
LinkChunk size
Next, we'll access various chunk sizes. Smaller chunks (but not too small!) are able to encapsulate atomic concepts which yields more precise retrieval. While larger chunks are more susceptible to noise. Popular strategies include using small chunks but retrieving a bit of the surrounding chunks around it (since it may have relevant info) or store multiple embeddings per document (ex. summary embedding per document).
1chunk_sizes = [100, 300, 500, 700, 900]
2for chunk_size in chunk_sizes:
3 experiment_name = f"chunk-size-{chunk_size}"
4 run_experiment(...)
5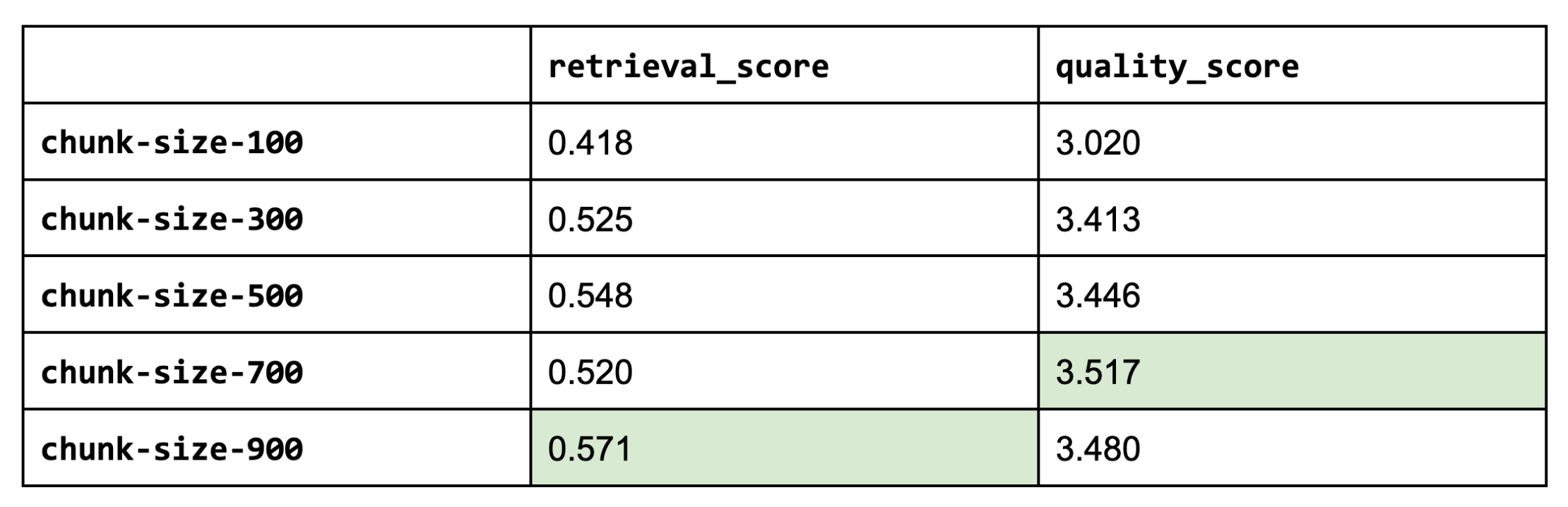
It appears that larger chunk sizes do help but tapers off (too much context might be too noisy). Larger chunk sizes aren’t always better.
Note: If we were to use larger chunk sizes (ours is based on characters), keep in mind that most open source embedding models have a maximum sequence length of 512 sub-word tokens. This means that if our chunk contains more than 512 sub-word tokens (4 chars ≈ 1 token), the embedding wouldn't account for it anyway (unless we fine-tune our embedding model to have longer sequence lengths).
1CHUNK_SIZE = 700
2CHUNK_OVERLAP = 50
3LinkNumber of chunks
Next, we'll experiment with the number of chunks to use. More chunks will allow us to add more context but too many could potentially introduce a lot of noise.
Note: The chunk_size we chose multiplied by the num_chunks needs to fit inside our LLM's context length. We're experimenting with the chunk size and number of chunks as if they were independent variables but they are heavily related. Especially since all of our LLMs have a finite maximum context length. So ideally, we would tune for a combination if chunk_size * num_chunks.
1num_chunks_list = [1, 3, 5, 7, 9]
2for num_chunks in num_chunks_list:
3 experiment_name = f"num-chunks-{num_chunks}"
4 run_experiment(...)
5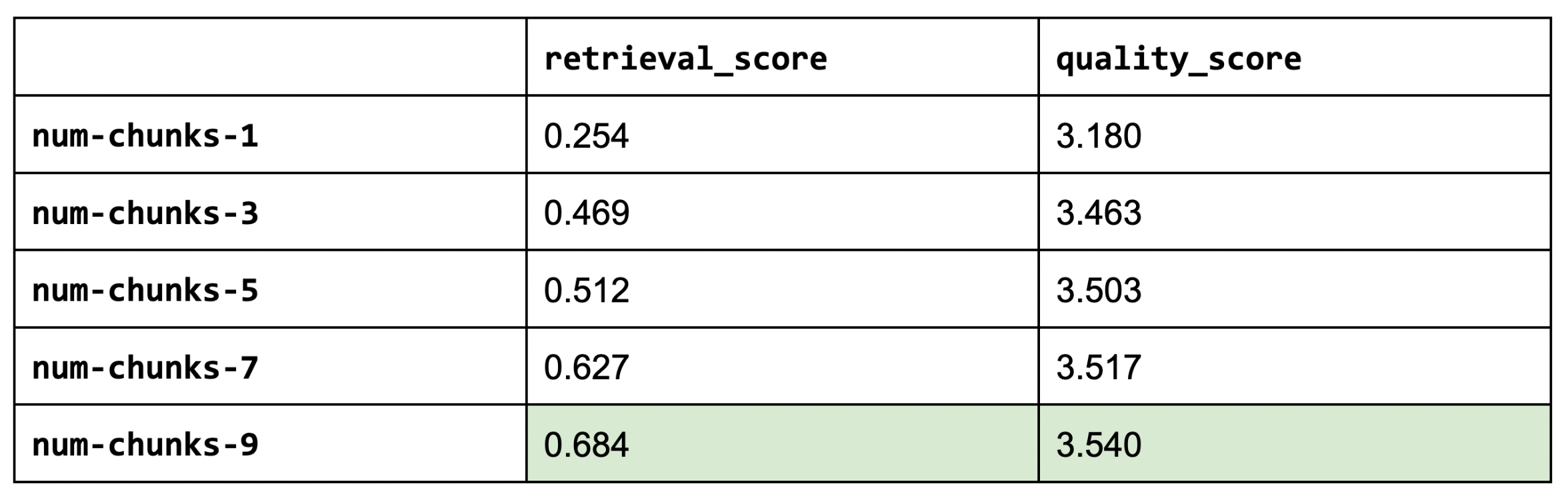

Increasing our number of chunks improves our retrieval and quality scores. We had to stop testing at num_chunks of 9 because we started to hit maximum context length often. This is a compelling reason to invest in extending context size via RoPE scaling (rotary position embeddings), etc.
Sanity check: Our retrieval score (in general) should increase as we increase the number of chunks.
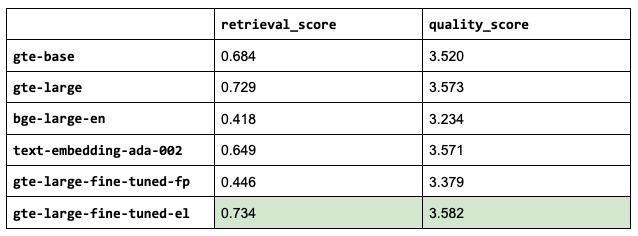
1NUM_CHUNKS = 9LinkEmbedding models
So far, we've used thenlper/gte-base as our embedding model because it's a relatively small (0.22 GB) and performant option. But now, let's explore other popular options such as thenlper/gte-large (0.67 GB), the current leader on the MTEB leaderboard, BAAI/bge-large-en (1.34 GB), and OpenAI's text-embedding-ada-002.
1embedding_model_names = ["thenlper/gte-base", "thenlper/gte-large", "BAAI/bge-large-en", "text-embedding-ada-002"]
2for embedding_model_name in embedding_model_names:
3 experiment_name = f"{embedding_model_name.split('/')[-1]}"
4 run_experiment(...)
5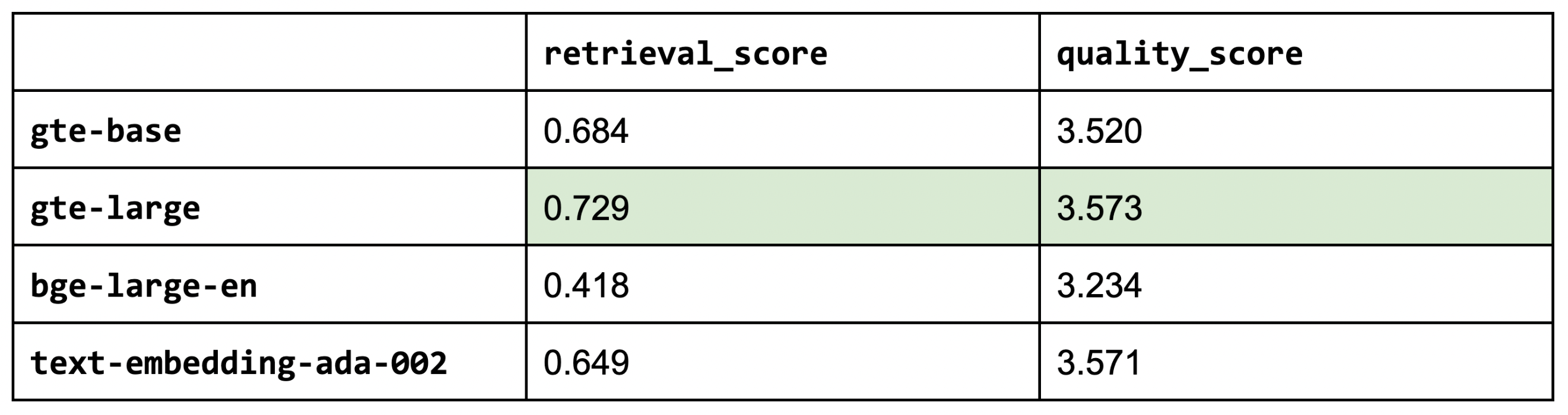
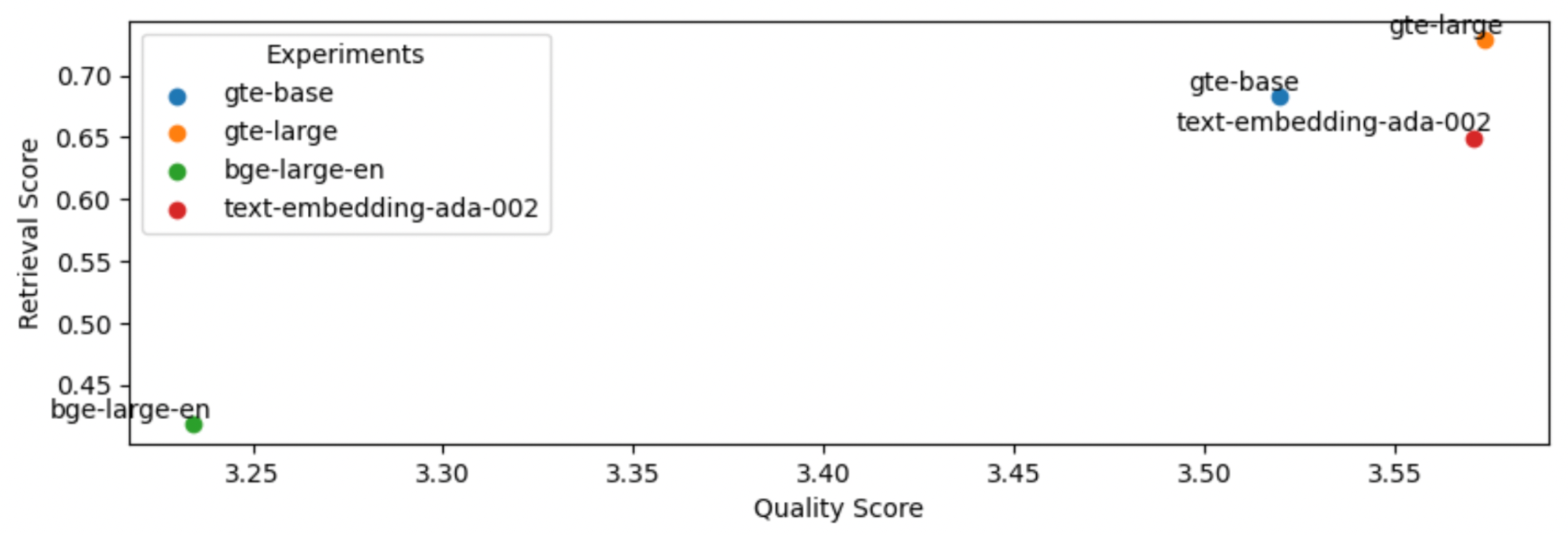
This is an interesting outcome because the #1 (BAAI/bge-large-en) on the current leaderboard isn't necessarily the best for our specific task. Using the smaller thenlper/gte-large produced the best retrieval and quality scores in our experiments.
1EMBEDDING_MODEL_NAME = "thenlper/gte-large"LinkOSS vs. closed LLMs
We're now going to use the best configurations from above to evaluate different choices for the main LLM.
Note:
We've been using a specific LLM so far to decide on the configuration so that specific LLM's performance here will be a bit biased.
This list is not exhaustive and even for the LLMs we use, there are versions with longer context windows available.
1llms = ["gpt-3.5-turbo",
2 "gpt-4",
3 "gpt-4-1106-preview",
4 "meta-llama/Llama-2-7b-chat-hf",
5 "meta-llama/Llama-2-13b-chat-hf",
6 "meta-llama/Llama-2-70b-chat-hf",
7 "codellama/CodeLlama-34b-Instruct-hf",
8 "mistralai/Mistral-7B-Instruct-v0.1",
9 "mistralai/Mixtral-8x7B-Instruct-v0.1"]
10for llm in llms:
11 experiment_name = f"{llm.split('/')[-1].lower()}"
12 run_experiment(...)
13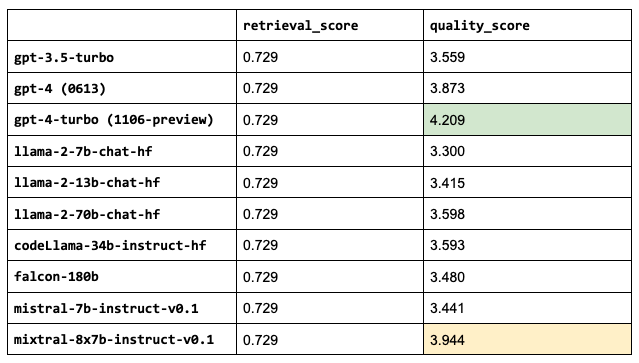
Sanity check: the retrieval scores are all the same because the LLM we choose doesn’t impact that part of our application.mixtral-8x7b-instruct-v0.1 outperforms the other OSS LLMs and even the current gpt-4 (currently 0613) and not too far behind gpt-4-turbo (currently 1106-preview).
1LLM = "mistralai/Mixtral-8x7B-Instruct-v0.1"Note: Some of our LLMs have much larger context lengths, ex. gpt-4 is 8,192 tokens and gpt-3.5-turbo-16k is 16,384 tokens. We could increase the number of chunks that we use for these since we saw that increasing num_chunks continued to improve the retrieval and quality scores. However, we will keep this value fixed for now since the performance started to taper off anyway and so we can compare these performances under the exact same configurations.
LinkMoEs without context
Curious how well these mixture of experts (MoE) fare without any context.
1moes = ["gpt-4",
2 "gpt-4-1106-preview",
3 "mistralai/Mixtral-8x7B-Instruct-v0.1"]
4for moe in moes:
5 experiment_name = f"without-context-{moe.split('/')[-1].lower()}"
6 run_experiment(num_chunks=0, ...)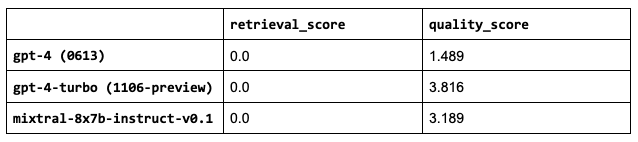
It seems that retrieving context is still very helpful even with a MoE architectures and with more recent training data cutoff dates. However, what makes the smaller mixtral model out perform gpt-4-0613 is to be determined.
LinkFine-tuning
Everything we have explored so far involves optimizing for how our data is preprocessed and using our models (embedding, LLM, etc.) as is. However, it's also worth exploring fine-tuning our models with data unique to our use case. This could help us better represent our data and ultimately increase our retrieval and quality scores. In this section, we're going to fine-tune our embedding model. The intuition here is that it may be worth it to learn a more contextual representation of our tokens than the default embedding models can. This can especially be impactful if we have a lot of:
new tokens that the default tokenization process creates subtokens out of that lose the significance of the token
existing tokens that have contextually different meanings in our use case
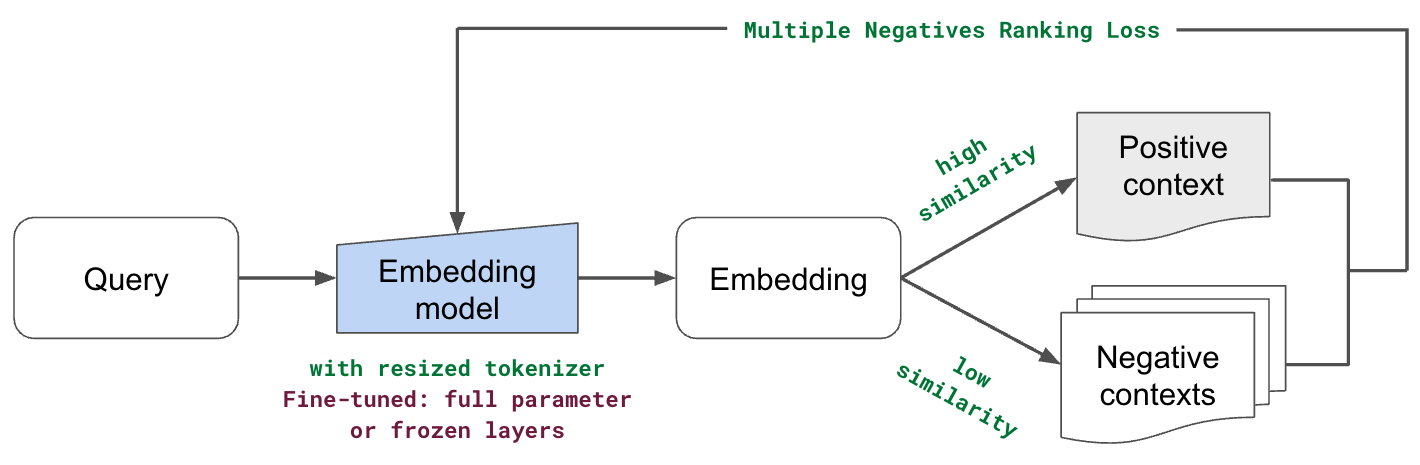
When it comes to fine-tuning our embedding model, we will exploring two approaches:
full parameter: including the embedding layer and all subsequent encoder layers (transformer blocks)
embedding layer: to better represent our unique subtokens and avoid overfitting (version of linear adapter)
Note: we will not be be exploring fine-tuning our LLM in this section because our previous experiments (LoRa vs. full parameter) have shown that fine-tuning has helped tremendously with form not facts, which in our case won't help too much (compared to for ex. SQL generation). However, your use cases might benefit from fine-tuning, so be sure to check out our Anyscale Endpoints fine-tuning to easily tune and serve models (fully hosted or private on your cloud).
LinkSynthetic dataset
Our first step will be to create a dataset to fine-tune our embedding model on. Our current embedding models have been trained via self-supervised learning (word2vec, GloVe, next/masked token prediction, etc.) and so we will continue fine-tuning with a self-supervised workflow. We're going to reuse a very similar approach as our cold start QA dataset section earlier so that we can map sections in our data to questions. The fine-tuning task here will be for the model to determine which sections in our dataset maps best to the input query. This optimization task will allow our embedding model to learn better representations of tokens in our dataset.
Note: While we could create a dataset mapping section titles with section text, we are creating a synthetic Q&A dataset because it will be most representative of the types of data we want to learn how to embed.
Our prompt is going to be a bit different because we want to generate a variety of different questions and we're going to use llama-70b here so that we can scale this QA generation process (and avoid any rate limits). To be thorough, we're going to generate one question from every section in our dataset so that we can try to capture as many unique tokens as possible.
1system_content = f"""
2Create one question using only the context provided starting with "What", "How" or "Why". Only respond with the question, don't say anything else (unecessary starting words, hints, etc.)
3"""1# Generate questions
2embedding_qa = []
3sections = sections_ds.take_all()
4max_context_length = int(0.5*MAX_CONTEXT_LENGTHS[LLM]-get_num_tokens(system_content))
5for section in tqdm(sections):
6 user_content = trim(
7 text=f"context: {section['text']}",
8 max_context_length=max_context_length)
9 response = generate_response(
10 llm="meta-llama/Llama-2-70b-chat-hf",
11 temperature=0.0,
12 stream=False,
13 system_content=system_content,
14 user_content=user_content,
15 max_retries=1)
16 if response:
17 embedding_qa.append({"question": response, "source": section["source"]})
18print (len(embedding_qa))LinkTraining data
We're now going to split our dataset into training and validation splits.
1from sentence_transformers import InputExample
2
3# Split counts
4num_train_samples = int(len(embedding_qa)*0.8)
5emb_qa_train = embedding_qa[:num_train_samples]
6emb_qa_val = embedding_qa[num_train_samples:]
7
8# Training dataset
9train_dataset = []
10for item in tqdm(emb_qa_train):
11 query = item["question"]
12 source_text = fetch_text(item["source"])
13 example = InputExample(texts=[query, source_text])
14 train_dataset.append(example)
15LinkValidation
Our validation evaluation criteria involves an information retrieval (IR) evaluator that will retrieve the top k similar documents from the corpus for each query. The InformationRetrievalEvaluator requires the following inputs:
queries:
Dict[str, str]# qid => querycorpus:
Dict[str, str]# cid => docrelevant_docs:
Dict[str, Set[str]]# qid => Set[cid]
Note: While our dataset may have multiple valid sections for a particular query, we will treat all other sections besides the one used to generate the query, as negative samples. This isn't an ideal scenario but the noise introduced is minimal, especially since we are using this to tune a representation layer (and not for a classification task).
1from sentence_transformers.evaluation import InformationRetrievalEvaluator
2
3# Validation dataset
4queries, corpus, relevant_docs = {}, {}, {}
5for i, item in tqdm(enumerate(emb_qa_val), total=len(emb_qa_val)):
6 queries[f"qid_{i}"] = item["question"]
7 corpus[f"cid_{i}"] = fetch_text(item["source"])
8 relevant_docs[f"qid_{i}"] = set([f"cid_{i}"])
9evaluator = InformationRetrievalEvaluator(queries, corpus, relevant_docs)We'll be using MultipleNegativesRankingLoss as our loss function. It will use the data points (InputExample(texts=[query, source_text]) in our training data as positive pairs and all other combinations as negative pairs. And the objective will be to increase the cosine similarity (default similarity_fct) for our positive pair and decrease it for the other pairs.
LinkEmbedding model
Now we're ready to initialize our embedding model for fine-tuning.
1from sentence_transformers import SentenceTransformer
2embedding_model = SentenceTransformer(EMBEDDING_MODEL_NAME) # gte-largeSentenceTransformer(
(0): Transformer({'max_seq_length': 512, 'do_lower_case': False}) with Transformer model: BertModel
(1): Pooling({'word_embedding_dimension': 1024, 'pooling_mode_cls_token': False, 'pooling_mode_mean_tokens': True, 'pooling_mode_max_tokens': False, 'pooling_mode_mean_sqrt_len_tokens': False})
(2): Normalize()
)
LinkResize tokenizer
While our tokenizer can represent new subtokens that are part of the vocabulary, it might be very helpful to explicitly add new tokens to our base model (BertModel) in our cast to our transformer. And then we can use resize_token_embeddings to adjust the model's embedding layer prior to fine-tuning. This can be very useful for contextual use cases, especially if many tokens are new or existing tokens have a very different meaning in our context.
1def get_unique_words(texts):
2 all_text = " ".join(texts) # join all texts
3 all_text = all_text.replace("_", " ") # replace underscores (ex. variable names)
4 words = re.findall(r'\b[a-zA-Z]+\b', all_text) # only letters
5 words = [word.lower() for word in words] # lower
6 return set(words)1# Get tokens that are OOV (out of vocabulary)
2new_words = []
3vocab = embedding_model.tokenizer.get_vocab().keys()
4texts = [section["text"] for section in sections_ds.take_all()]
5unique_words = get_unique_words(texts=texts)
6for word in tqdm(unique_words):
7 if word not in vocab:
8 new_words.append(word)
9
10# Inspect
11print (len(new_words))
12print (new_words[:10])5790
['dilation', 'azurealiyunvsphere', 'rlmoduleconfig', 'multipledispatch', 'specifying', 'pycaret', 'duelingqmodel', 'callable', 'autoscaling', 'iterators']
Now we can add these new words to our tokenizer and they won’t be split into subtokens:
1# Add new words to tokenizer
2print (len(embedding_model.tokenizer))
3embedding_model.tokenizer.add_tokens(new_words)
4print (len(embedding_model.tokenizer))
5
6# Resize tokenizer
7print (embedding_model._modules["0"]._modules["auto_model"]._modules["embeddings"]._modules["word_embeddings"])
8embedding_model._modules["0"]._modules["auto_model"].resize_token_embeddings(len(embedding_model.tokenizer))
9embedding_model._modules["0"]._modules["auto_model"]._modules["embeddings"]._modules["word_embeddings"].padding_idx = 0
10print (embedding_model._modules["0"]._modules["auto_model"]._modules["embeddings"]._modules["word_embeddings"])Embedding(30522, 1024, padding_idx=0)
Embedding(36312, 1024, padding_idx=0)
LinkFull parameter
Our full parameter fine-tuning approach will tune all of the following weights:
BertModel(
(embeddings): BertEmbeddings,
(encoder): BertEncoder
(pooler): BertPooler)
1from sentence_transformers.losses import MultipleNegativesRankingLoss
2from torch.utils.data import DataLoader
3
4# Training setup
5num_epochs = 2
6batch_size = 4
7train_dataloader = DataLoader(train_dataset, batch_size=batch_size)
8loss = MultipleNegativesRankingLoss(embedding_model) # MNR Loss
9warmup_steps = int(0.1 * num_epochs * len(train_dataloader)) # not used
10
11# Train
12experiment_name = "gte-large-fine-tuned-fp"
13gte_large_ft_path = str(Path(EFS_DIR, experiment_name))
14embedding_model.fit(
15 train_objectives=[(train_dataloader, loss)],
16 epochs=num_epochs,
17 warmup_steps=0,
18 optimizer_params={"lr": 1e-8},
19 weight_decay=0,
20 output_path=gte_large_ft_path,
21 show_progress_bar=True,
22 evaluator=evaluator,
23 callback=val_callback)
24EPOCH: 0, VAL SCORE:0.5242
EPOCH: 1, VAL SCORE:0.52
Now we're ready to actually apply this fine-tuned embedding model on our test evaluation dataset. We can simply pass in our model artifact directory for the embedding_model_name because HuggingFaceEmbeddings accepts a string that can be either a directory or the model's name. If a directory matches with the input string, then it will load the model from that location first before trying to search on HF's hub.
1sql_dump_fp = Path(EFS_DIR, "sql_dumps", f"{experiment_name}_{CHUNK_SIZE}_{CHUNK_OVERLAP}.sql")
2run_experiment(sql_dump_fp, **kwargs)gte-large-fine-tuned-fp
retrieval score: 0.4463276836158192
quality score: 3.378531073446328
This didn't really improve our overall application's retrieval or quality score. This doesn't necessarily mean that fine-tuning is not useful but might not always be worth the effort.
synthetic data is not exactly like the types of questions that users ask (might be worth creating a dataset of more realistic queries or prompt tuning for more synthetic data that is more representative of user queries).
Fine-tuning the entire embedding model on our small embedding dataset might be causing overfitting.
Our experiment's evaluation is on a small dataset so slightly tuning embeddings via MNR may not increase retrieval recall much/if at all.
LinkEmbedding layer
To help mitigate the overfitting, we can avoid retraining the entire embedding model and freeze all layers except for the embedding layer (word/subtoken embedding only, not the positional or token type layers).
BertEmbeddings(
(word_embeddings): Embedding(30522, 1024, padding_idx=0)
(position_embeddings): Embedding(512, 1024)
(token_type_embeddings): Embedding(2, 1024)
(LayerNorm): LayerNorm((1024,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
1# Reinitialize base embedding model
2embedding_model = SentenceTransformer(EMBEDDING_MODEL_NAME) # gte-large
3
4# Unfreeze embedding layers
5for param in embedding_model._modules["0"]._modules["auto_model"]._modules["embeddings"].parameters(): param.requires_grad = True
6
7# Freeze Bert encoder layers
8for param in embedding_model._modules["0"]._modules["auto_model"]._modules["encoder"].parameters(): param.requires_grad = FalseNow we can run the exact same training workflow as we did with full parameter fine-tuning:
1# Training setup
2num_epochs = 2
3batch_size = 4
4train_dataloader = DataLoader(train_dataset, batch_size=batch_size)
5loss = MultipleNegativesRankingLoss(embedding_model)
6warmup_steps = int(0.1 * num_epochs * len(train_dataloader)) # not used
7
8# Train
9experiment_name = "gte-large-fine-tuned-el"
10gte_large_ft_path = str(Path(EFS_DIR, experiment_name))
11embedding_model.fit(
12 train_objectives=[(train_dataloader, loss)],
13 epochs=num_epochs,
14 warmup_steps=0,
15 optimizer_params={"lr": 1e-5},
16 weight_decay=0,
17 output_path=gte_large_ft_path,
18 show_progress_bar=True,
19 evaluator=evaluator,
20 callback=val_callback)EPOCH: 0, VAL SCORE:0.7938
EPOCH: 1, VAL SCORE:0.7965
1sql_dump_fp = Path(EFS_DIR, "sql_dumps", f"{experiment_name}_{CHUNK_SIZE}_{CHUNK_OVERLAP}.sql")
2run_experiment(sql_dump_fp, **kwargs)gte-large-fine-tuned-el
retrieval score: 0.7344632768361582
quality score: 3.5819209039548023
Much better validation scores and overall better performance but it's not worth the effort compared to using our base gte-large embedding model. This again can be improved with larger/higher quality datasets and perhaps even a larger testing dataset to capture small improvements in our retrieval scores.
Note: even though the retrieval scores are the same, the quality scores differ due to the order in which the new embedding models determine the top k relevant chunks and if different relevant sources were introduced.
1experiment_name = "gte-large-fine-tuned-el"
2EMBEDDING_MODEL_PATH = str(Path(EFS_DIR, experiment_name)) # can pass this in directly for embedding_model_name
3SQL_DUMP_FP = Path(EFS_DIR, "sql_dumps", f"{experiment_name}_{CHUNK_SIZE}_{CHUNK_OVERLAP}.sql")LinkPrompt engineering
There's too much we can do when it comes to engineering the prompt (x-of-thought, multimodal, self-refine, query decomposition, etc.) so we're going to try out just a few interesting ideas. We're going to allow the LLM to ignore anything not relevant. The idea here is to show how quickly we can go from prompt engineering to evaluation report.
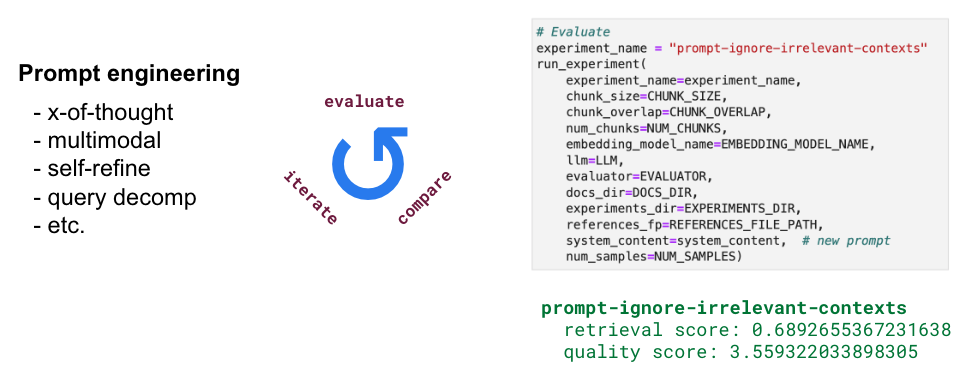
1# Prompt
2generation_system_content = "Answer the query using the context provided. Be succinct. Contexts are organized in a list of dictionaries [{'text': <context>}, {'text': <context>}, ...]. Feel free to ignore any contexts in the list that don't seem relevant to the query. "
3
4# Evaluate
5experiment_name = "prompt-ignore-contexts"
6run_experiment(
7 experiment_name=experiment_name,
8 generation_system_content=generation_system_content, # new prompt
9 **kwargs)prompt-ignore-contexts
retrieval score: 0.7288135593220338
quality score: 3.519774011299435
It seems this specific prompt engineering effort didn't help improve the quality of our system. As we mentioned earlier, there are too many other ways we can engineer our prompt and we encourage you to explore more. What’s important here is that we have a clean and simple way to evaluate anything that we want to experiment with. However, we have empirically found that improving the quality of our retrieval system and the data flywheel (where we fix our documentation itself) has had a much larger impact on the overall quality of our system.
1SYSTEM_CONTENT = "Answer the query using the context provided. Be succinct."LinkLexical search
We're going to now supplement our vector embedding based search with traditional lexical search, which searches for exact token matches between our query and document chunks. Our intuition here is that lexical search can help identify chunks with exact keyword matches where semantic representation may fail to capture. Especially for tokens that are out-of-vocabulary (and so represented via subtokens) with our embedding model. But our embeddings based approach is still very advantageous for capturing implicit meaning, and so we're going to combine several retrieval chunks from both vector embeddings based search and lexical search.
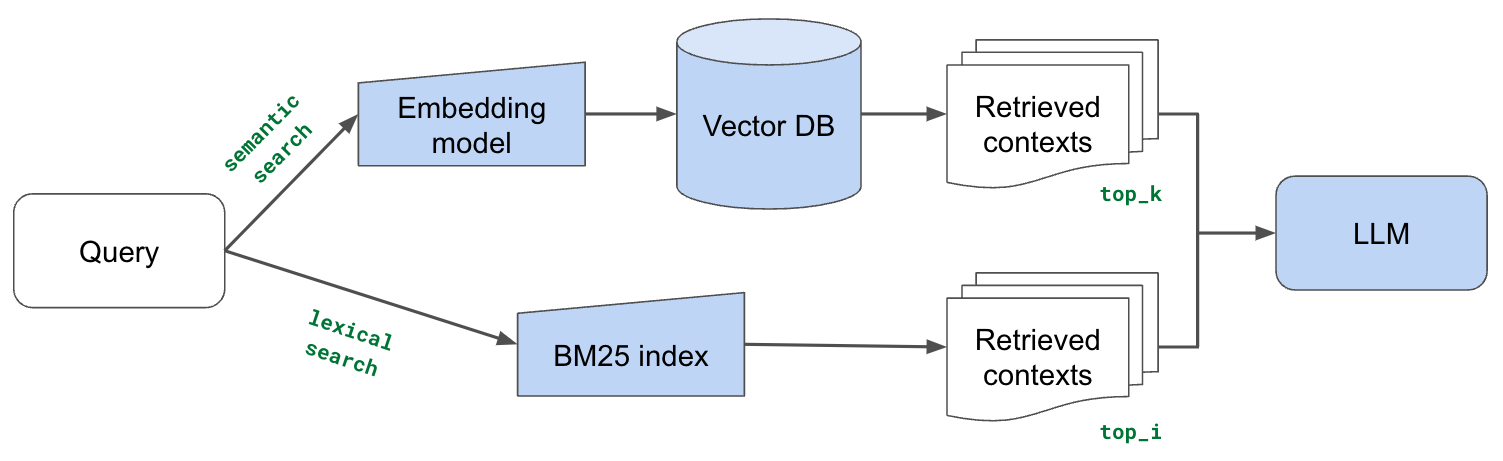
LinkBM25
Let's apply lexical search using BM25, which is a ranking algorithm that rewards unique token matches between our query and contexts.
1import re
2from rank_bm25 import BM25Okapi
3
4# BM25 index
5texts = [re.sub(r"[^a-zA-Z0-9]", " ", chunk[1]).lower().split() for chunk in chunks]
6lexical_index = BM25Okapi(texts)Similar to our semantic_search function to retrieve the relevant context, we can implement a search function to use our lexical index to retrieve relevant context.
1def lexical_search(index, query, chunks, k):
2 query_tokens = query.lower().split() # preprocess query
3 scores = index.get_scores(query_tokens) # get best matching (BM) scores
4 indices = sorted(range(len(scores)), key=lambda i: -scores[i])[:k] # sort and get top k
5 lexical_context = [{
6 "id": chunks[i][0],
7 "text": chunks[i][1],
8 "source": chunks[i][2],
9 "score": scores[i]} for i in indices]
10 return lexical_context
11
12# Retrieve top-k docs
13k = 3
14query = "What is the default batch size for map_batches?"
15top_docs = lexical_search(lexical_index, query, chunks, k=k)
16for item in top_docs:
17 print (item["source"])
18 print (item["text"])
19 print ()Transforming Data — Ray 2.7.1
Configuring batch size#
The default batch size depends on your resource type. If you’re using CPUs,
the default batch size is 4096. If you’re using GPUs, you must specify an explicit batch size.
LinkSemantic
Comparing this with the retrieved sources with our existing vector embedding based search shows that the two approaches, while different, both retrieved relevant sources. So, we're going to combine both approaches and feed it into the context for our LLM for generation.
ray.data.Dataset.map_batches — Ray 2.7.1
to a given map task. Default batch_size is 4096 with “default”.
compute – Either “tasks” (default) to use Ray Tasks or an
ActorPoolStrategy to use an autoscaling actor pool.
batch_format – If "default" or "numpy", batches are
Dict[str, numpy.ndarray].
LinkLexical experiments
Now let's incorporate this into our retrieval workflow by adding it to our generate.py/QueryAgent class. The main change will be to include the additional sources from lexical search:
1def QueryAgent():
2 def __init__(use_lexical_search=True, chunks=[...], **kwargs):
3 # Lexical search
4 self.chunks = chunks
5 self.lexical_index = None
6 if use_lexical_search:
7 texts = [re.sub(r"[^a-zA-Z0-9]", " ", chunk[1]).lower().split() for chunk in chunks]
8 self.lexical_index = BM25Okapi(texts)
9
10 def __call__(lexical_search_k=1, **kwargs):
11 # Add lexical search results
12 if self.lexical_index:
13 lexical_context = lexical_search(
14 index=self.lexical_index, query=query, chunks=self.chunks, k=lexical_search_k)
15 # Insert after <lexical_search_k> worth of semantic results
16 context_results[lexical_search_k:lexical_search_k] = lexical_contextAnd now we can run our experiment:
1lexical_search_k_list = [1, 3, 5]
2use_lexical_search = True
3for lexical_search_k in lexical_search_k_list:
4 experiment_name = f"lexical-search-bm25-{lexical_search_k}"
5 experiment_names.append(experiment_name)
6 run_experiment(
7 experiment_name=experiment_name,
8 use_lexical_search=use_lexical_search,
9 lexical_search_k=lexical_search_k,
10 **kwargs)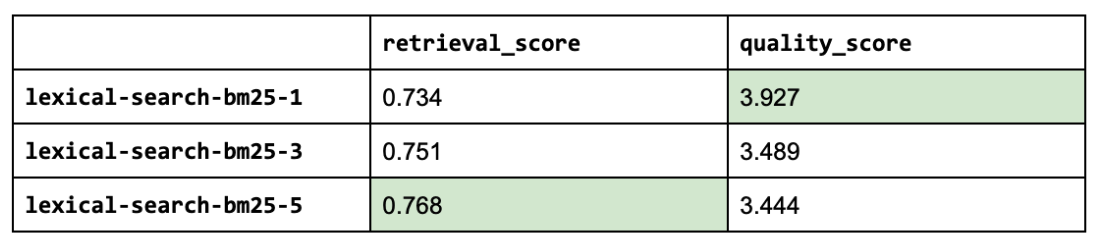
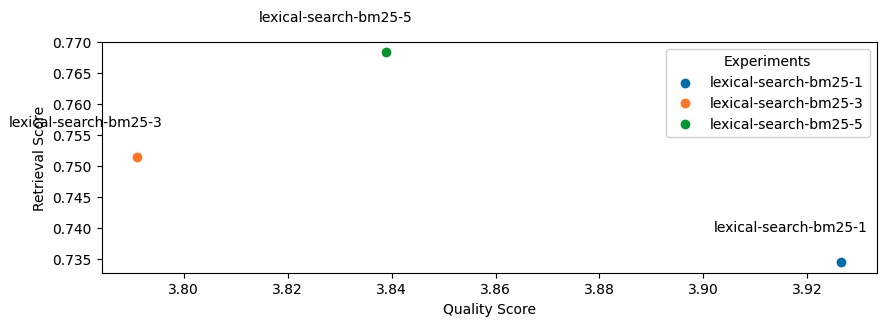
Seems like adding lexical search was not as impactful as we had hoped but this was just one aspect (keyword matching) of lexical search that we explored but there are many other useful features such as filtering, counts, etc. It's also worth exploring how we combine the lexical search results with semantic search results.
1USE_LEXICAL_SEARCH = False
2LEXICAL_SEARCH_K = 0LinkReranking
So far with all of our approaches, we've used an embedding model (+ lexical search) to identify the top k relevant chunks in our dataset. The number of chunks (k) has been a small number because we found that adding too many chunks did not help and our LLMs have restricted context lengths. However, this was all under the assumption that the top k retrieved chunks were truly the most relevant chunks and that their order was correct as well. What if increasing the number of chunks didn't help because some relevant chunks were much lower in the ordered list. And, semantic representations, while very rich, were not trained for this specific task.
In this section, we implement reranking so that we can use our semantic and lexical search methods to cast a much wider net over our dataset (retrieve many chunks) and then rerank the order based on the user's query. The intuition here is that we can account for gaps in our semantic representations with ranking specific to our use case. We'll train a supervised model that predicts which part of our documentation is most relevant for a given user's query. We'll use this prediction to then rerank the relevant chunks so that chunks from this part of our documentation are moved to the top of the list.
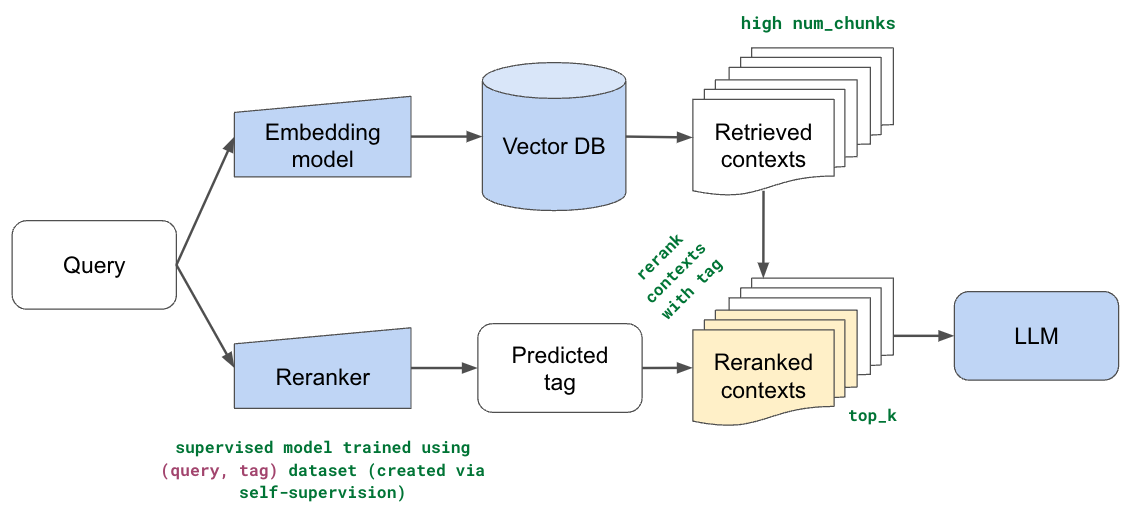
LinkDataset
We're going to reuse the QA dataset we created in our fine-tuning section because that dataset has questions that map with specific sections. We’ll create a feature called text that will concatenate the section title and the question. And we’ll use this feature as the input to our model to predict the appropriate. We add the section title (even though this information won’t be available during inference from our users queries) so that our model can learn how to represent key tokens that will be in the user’s queries.
1def get_tag(url):
2 return re.findall(r"docs\.ray\.io/en/master/([^/]+)", url)[0].split("#")[0]
3
4# Load data
5from pathlib import Path
6df = pd.read_json(Path(ROOT_DIR, "datasets", "embedding_qa.json"))
7df["tag"] = df.source.map(get_tag)
8df["section"] = df.source.map(lambda source: source.split("/")[-1])
9df["text"] = df["section"] + " " + df["question"]
10df.sample(n=5)
1# Map only what we want to keep
2tags_to_keep = ["rllib", "tune", "train", "cluster", "ray-core", "data", "serve", "ray-observability"]
3df["tag"] = df.tag.apply(lambda x: x if x in tags_to_keep else "other")
4Counter(df.tag)Counter({'rllib': 1269,
'tune': 979,
'train': 697,
'cluster': 690,
'data': 652,
'ray-core': 557,
'other': 406,
'serve': 302,
'ray-observability': 175})
LinkPreprocessing
We'll start by creating some preprocessing functions to better represent our data. For example, our documentation has many variables that are camel cased (ex. RayDeepSpeedStrategy). When a tokenizer is used on this, we often lose the individual tokens that we know to be useful and, instead, random subtokens are created.
Note: we didn't omnisciently know to create these unique preprocessing functions! This is all a result of methodical iteration. We train a model → view incorrect data points → view how the data was represented (ex. subtokenization) → update preprocessing → iterate ↺
1import re
2from transformers import BertTokenizer
3
4def split_camel_case_in_sentences(sentences):
5 def split_camel_case_word(word):
6 return re.sub("([a-z0-9])([A-Z])", r"\1 \2", word)
7 processed_sentences = []
8 for sentence in sentences:
9 processed_words = []
10 for word in sentence.split():
11 processed_words.extend(split_camel_case_word(word).split())
12 processed_sentences.append(" ".join(processed_words))
13 return processed_sentences
14
15def preprocess(texts):
16 texts = [re.sub(r'(?<=\w)([?.,!])(?!\s)', r' \1', text) for text in texts]
17 texts = [text.replace("_", " ").replace("-", " ").replace("#", " ").replace(".html", "").replace(".", " ") for text in texts]
18 texts = split_camel_case_in_sentences(texts) # camelcase
19 texts = [tokenizer.tokenize(text) for text in texts] # subtokens
20 texts = [" ".join(word for word in text) for text in texts]
21 return texts
22
23print (preprocess(["RayDeepSpeedStrategy"]))
24print (preprocess(["What is the default batch_size for map_batches?"]))['ray deep speed strategy']
['what is the default batch size for map batch ##es ?']
LinkTraining
Now we’re going to train a simple logistic regression model that will predict the tag given the input text.
1from sklearn.feature_extraction.text import TfidfVectorizer
2from sklearn.linear_model import LogisticRegression
3from sklearn.pipeline import Pipeline
4from sklearn.preprocessing import FunctionTransformer
5
6# Train classifier
7from rag.rerank import preprocess # for pickle
8reranker = Pipeline([
9 ("preprocess", FunctionTransformer(preprocess)),
10 ("vectorizer", TfidfVectorizer(lowercase=True)),
11 ("classifier", LogisticRegression(multi_class="multinomial", solver="lbfgs"))
12])
13reranker.fit(train_df["text"].tolist(), train_df["tag"].tolist())Note: we also trained a BERT classifier and while performance was better than our logistic classifier, these large networks suffer from overconfidence and we can't use a threshold based approach as we do below. And without the threshold approach (where we only rerank when the reranker is truly confident), then the quality score of our application does not improve.
1# Inference
2question = "training with deepspeed"
3custom_predict([question], classifier=reranker)[0]'train'
We're now ready to evaluate our trained reranking model. We're going to use a custom prediction function that will predict “other” unless the probability of the highest class is above a certain threshold.
1def custom_predict(inputs, classifier, threshold=0.3, other_label="other"):
2 y_pred = []
3 for item in classifier.predict_proba(inputs):
4 prob = max(item)
5 index = item.argmax()
6 if prob >= threshold:
7 pred = classifier.classes_[index]
8 else:
9 pred = other_label
10 y_pred.append(pred)
11 return y_pred
12
13# Evaluation
14metrics = {}
15y_test = test_df["tag"]
16y_pred = custom_predict(inputs=test_df["question"], classifier=reranker)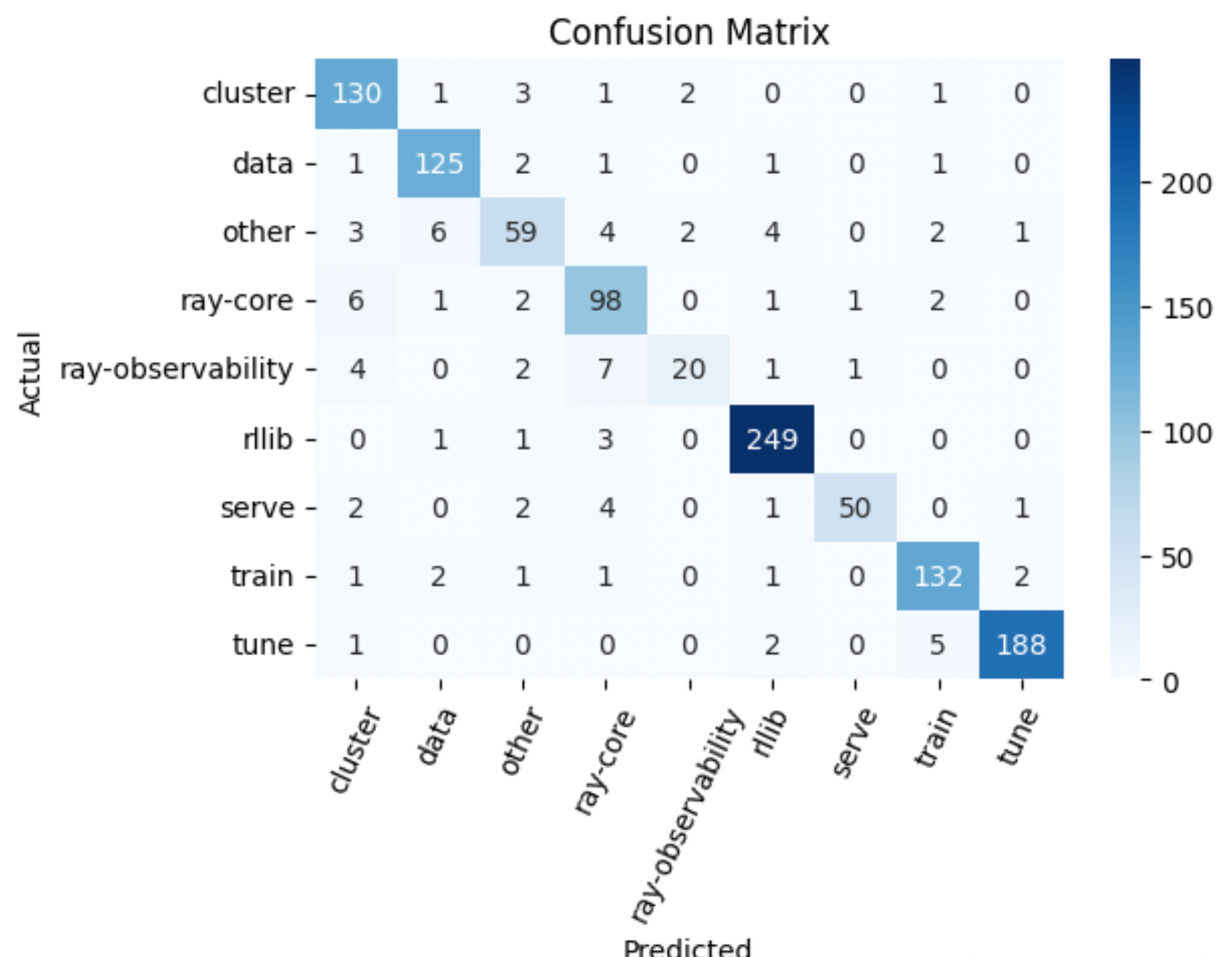
{
"precision": 0.9168129573272782,
"recall": 0.9171029668411868,
"f1": 0.9154520876579969,
"num_samples": 1146.0
}
LinkTesting
Besides just a metric based evaluation, we also want to assess how our model performs on some minimum functionality tests. We need all of these basic sanity checks to pass regardless of what type of model we use.
1# Basic tests
2tests = [
3 {"question": "How to train a train an LLM using DeepSpeed?", "tag": "train"},
4 ...
5 {"question": "How do I set a maximum episode length when training with Rllib", "tag": "rllib"}]
6for test in tests:
7 question = test["question"]
8 prediction = predict_proba(question=test["question"], classifier=reranker)[0][1]
9 print (f"[{prediction}]: {question} → {preprocess([question])}")
10 assert (prediction == test["tag"])[train]: How to train a train an LLM using DeepSpeed? → ['how to train a train an ll ##m using deep speed ?']
...
[rllib]: How do I set a maximum episode length when training with Rllib → ['how do i set a maximum episode length when training with r ##lli ##b']
LinkReranking experiments
Now we're ready to apply our reranking model post retrieval using these steps:
Increase the retrieved context (can experiment with this) so that we can apply reranking to yield a smaller subset (
num_chunks). The intuition here is that we'll use semantic and lexical search to retrieve N chunks (N > k) and then we'll use reranking to reorder the retrieved results (top k).If the predicted tag is above the
threshold, then we will move all retrieved sources from that tag to the top. If the predicted tag is below the threshold, then no reranking will be performed. The intuition here is that, unless we are confident about which parts of our documentation a specific query pertains to (or if it happens to involve multiple parts), then we will not incorrectly rerank the results.Perform generation using the top k retrieved chunks.
We're going to alter our QueryAgent class directly to include reranking:
1class QueryAgent():
2 def __init__(rerank=True, **kwargs):
3 # Reranker
4 self.reranker = None
5 if rerank:
6 reranker_fp = Path(EFS_DIR, "reranker.pkl")
7 with open(reranker_fp, "rb") as file:
8 self.reranker = pickle.load(file)
9
10 def __call__(rerank_threshold=0.3, rerank_k=7, **kwargs):
11 # Rerank
12 if self.reranker:
13 predicted_tag = custom_predict(
14 inputs=[query], classifier=self.reranker, threshold=rerank_threshold)[0]
15 if predicted_tag != "other":
16 sources = [item["source"] for item in context_results]
17 reranked_indices = get_reranked_indices(sources, predicted_tag)
18 context_results = [context_results[i] for i in reranked_indices]
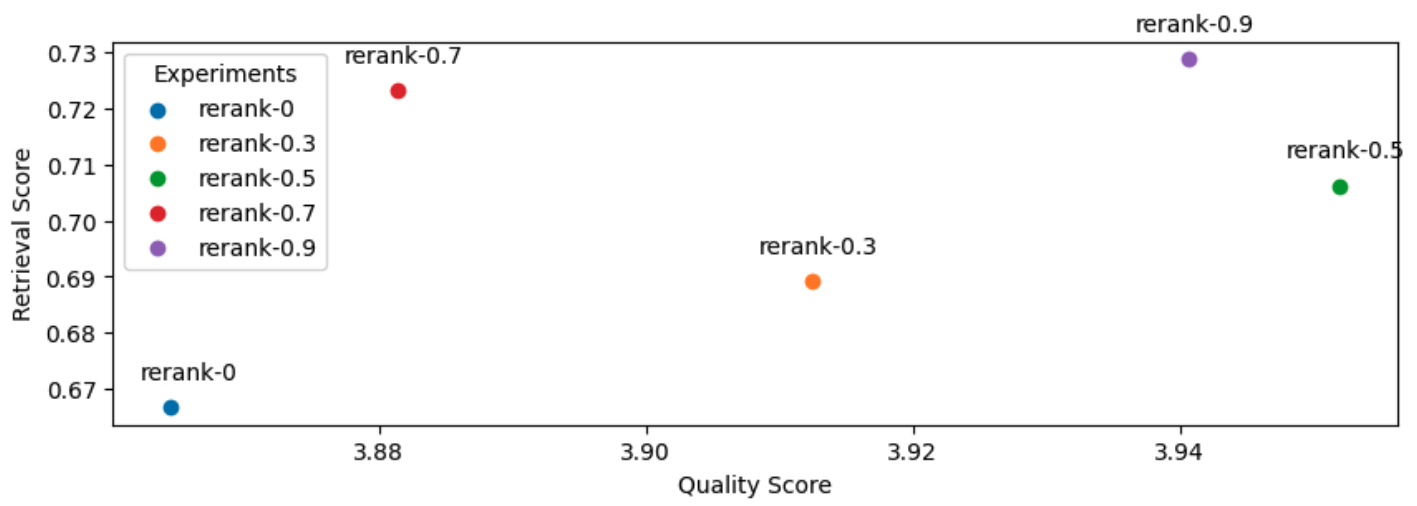
19 context_results = context_results[:rerank_k]And with that, let's use our query agent augmented with reranking on an evaluation run. We will experiment with various reranking threshold values. Note: a threshold of zero is the same as not using any threshold.
1# Experiment
2rerank_threshold_list = [0, 0.3, 0.5, 0.7, 0.9]
3use_reranking = True
4for rerank_threshold in rerank_threshold_list:
5 experiment_name = f"rerank-{rerank_threshold}"
6 experiment_names.append(experiment_name)
7 run_experiment(
8 experiment_name=experiment_name,
9 num_chunks=30, # increased num chunks since we will retrieve top k
10 rerank_k=NUM_CHUNKS + LEXICAL_SEARCH_K, # subset of larger num_chunks
11 **kwargs)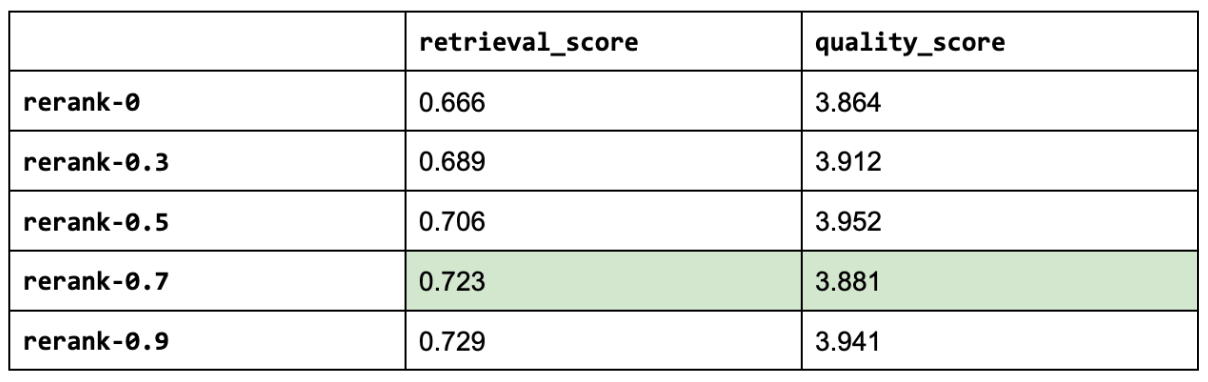
1original_num_chunks = NUM_CHUNKS
2NUM_CHUNKS = 30
3USE_RERANKING = True
4RERANK_THRESHOLD = 0.5
5RERANK_K = original_num_chunks + LEXICAL_SEARCH_KNote: there is still a lot more to experiment with reranking (increasing the initial num_chunks, adding lexical search results after reranking, weighted reranking where we promote the top N classes, etc.)
And as a reference, here are the top three experiments so far:
[('gpt-4-1106-preview',
{'retrieval_score': 0.7288135593220338, 'quality_score': 4.209039548022599}),
('rerank-0.5',
{'retrieval_score': 0.7062146892655368,
'quality_score': 3.9519774011299433}),
('prompt-ignore-contexts',
{'retrieval_score': 0.7344632768361582, 'quality_score': 3.943502824858757})]
LinkCost analysis
Besides just performance, we also want to evaluate the cost of our configurations (especially given the high price points of larger LLMs). We’re going to break this down into prompt and sampled pricing. The prompt size is the number of characters in our system, assistant and user contents (which includes the retrieved contexts). And the sampled size is the number of characters the LLM generated in its response.
Note: Our OSS models are served with Anyscale Endpoints.
1# Pricing per 1M tokens
2# Pricing per 1M tokens
3PRICING = {
4 "gpt-3.5-turbo": {
5 "prompt": 1.5,
6 "sampled": 2
7 },
8 "gpt-4": {
9 "prompt": 30,
10 "sampled": 60
11 },
12 "gpt-4-1106-preview": {
13 "prompt": 10,
14 "sampled": 30
15 },
16 "llama-2-7b-chat-hf": {
17 "prompt": 0.15,
18 "sampled": 0.15
19 },
20 "llama-2-13b-chat-hf": {
21 "prompt": 0.25,
22 "sampled": 0.25
23 },
24 "llama-2-70b-chat-hf": {
25 "prompt": 1,
26 "sampled": 1
27 },
28 "codellama-34b-instruct-hf": {
29 "prompt": 1,
30 "sampled": 1
31 },
32 "mistral-7b-instruct-v0.1": {
33 "prompt": 0.15,
34 "sampled": 0.15
35 },
36 "mixtral-8x7b-instruct-v0.1": {
37 "prompt": 0.50,
38 "sampled": 0.50
39 }
40}
41for llm in llms:
42 cost_analysis(llm=llm)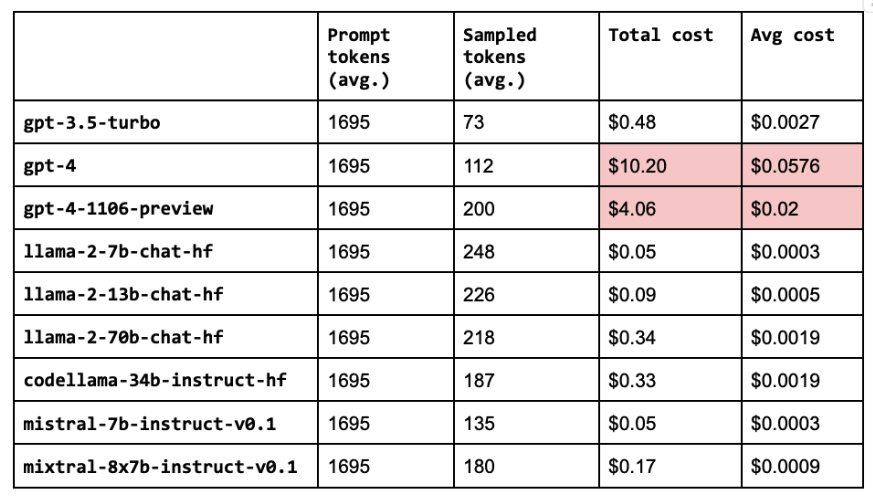
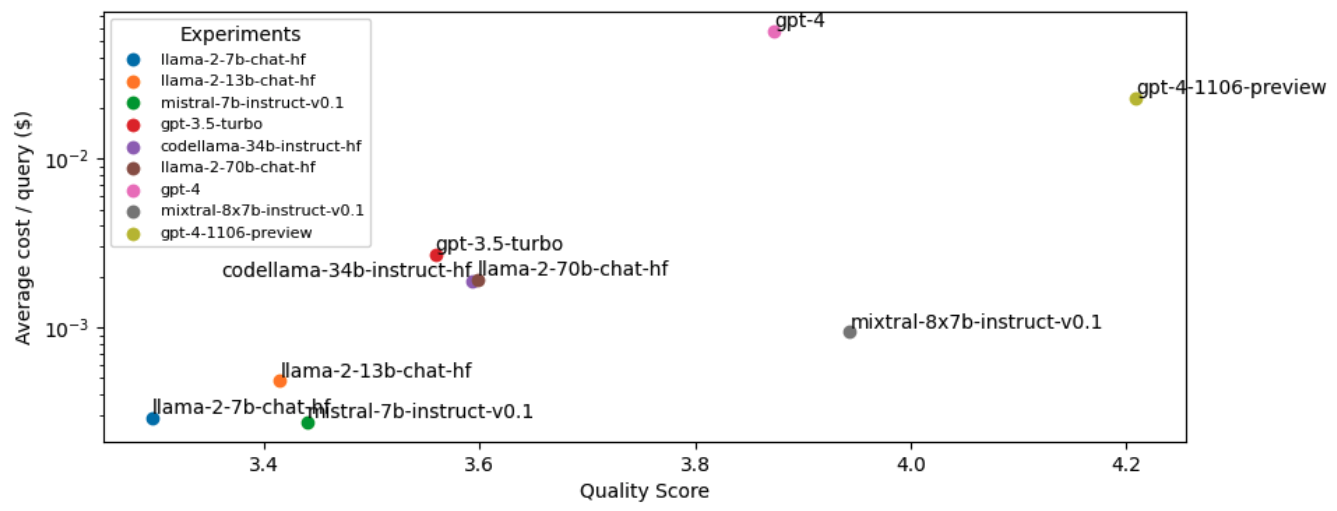
Note: This cost analysis is performed with our original experiments before lexical search, reranking, etc. since we haven't run experiments with these improvements on the other OSS and closed source LLMs yet.
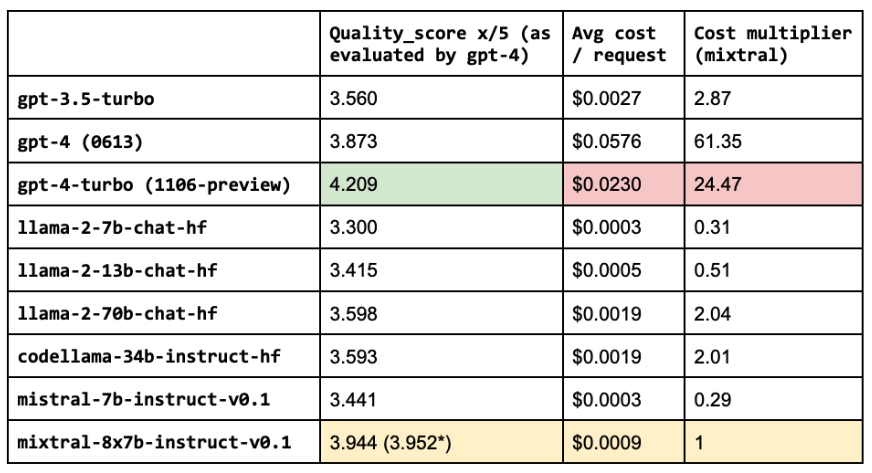
(*) quality score with fine-tuned embeddings, prompt engineering, lexical search, reranking, etc.
LinkRouting
It seems that the most performant LLM, gpt-4-turbo, is also very expensive. While our OSS LLM (mixtral-8x7b-instruct-v0.1) is very close in quality but ~25X more cost-effective. However, we want to be able to serve the most performant and cost-effective solution. We can close this gap in performance between open source and proprietary models by routing queries to the right LLM according to the complexity or topic of the query. For example, in our application, open source models perform really well on simple queries where the answer can be easily inferred from the retrieved context. However, the OSS models fall short for queries that involve reasoning, numbers or code examples. To identify the appropriate LLM to use, we can train a classifier that takes the query and routes it to the best LLM.
Question for gpt-4:
{'question': 'if I am inside of a anyscale cluster how do I get my cluster-env-build-id', 'target': 0}
Question for OSS LLM:
{'question': 'what is num_samples in tune?', 'target': 1}
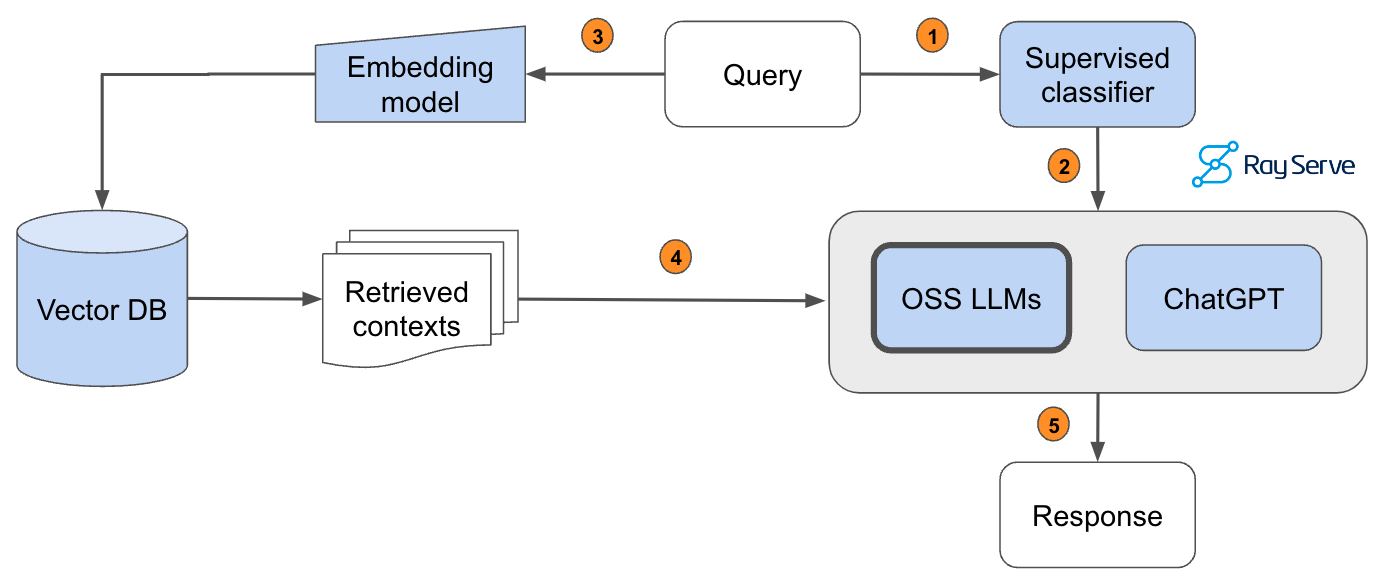
Pass the query to a supervised classifier that will determine which LLM is appropriate to answer it.The predicted LLM receives the query.Pass the query to our embedding model to semantically represent it.Pass the retrieved context to the predicted LLM.Generate the response.
In order to implement this, we hand-annotated a dataset of 1.8k queries according to which model (gpt-4 (label=0) or OSS LLM (label=1)) would be appropriate -- by default we route to OSS LLM and only if the query needs more advanced capabilities do we send the query to gpt-4. We then evaluate the performance of the model on a test dataset that has been scored with an evaluator.
1# Train classifier
2vectorizer = CountVectorizer()
3classifier = LogisticRegression(multi_class="multinomial", solver="lbfgs")
4router = Pipeline([("vectorizer", vectorizer), ("classifier", classifier)])
5router.fit(texts, labels)
6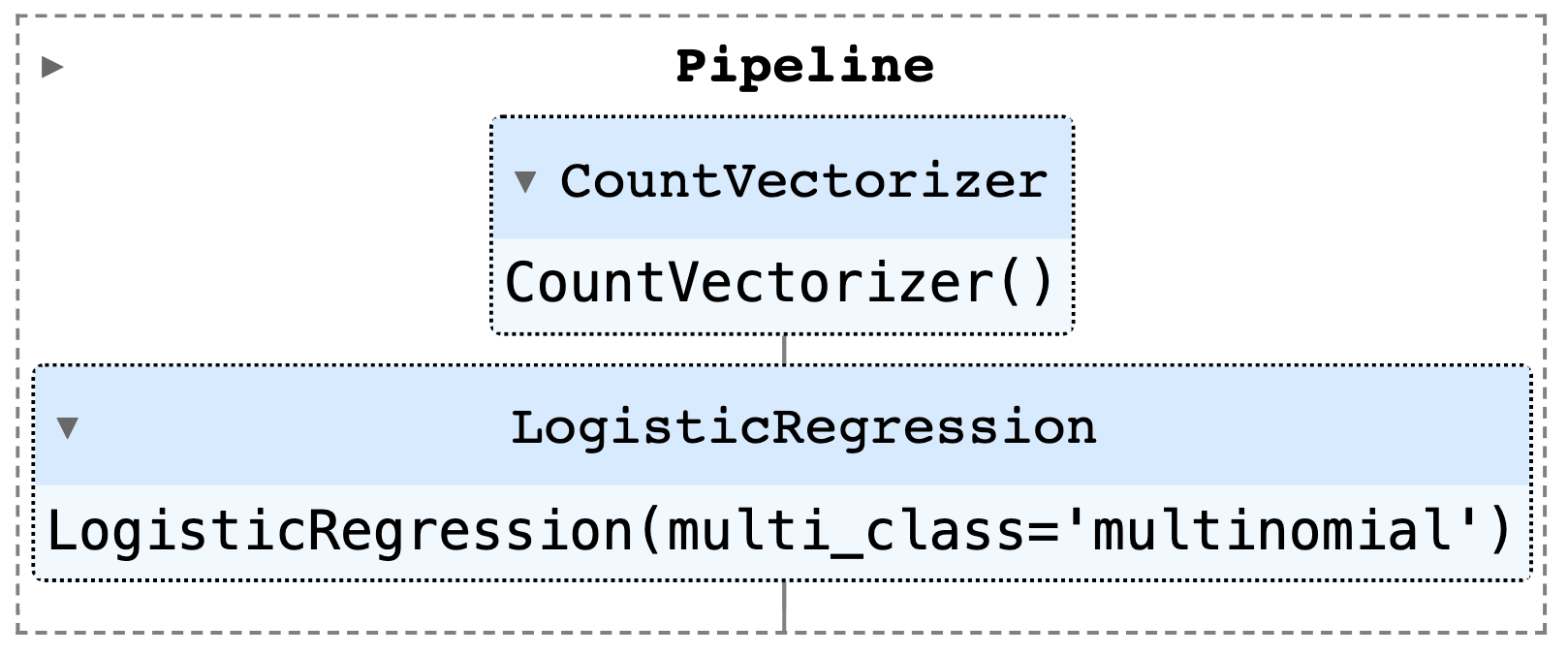
{
"precision": 0.9191264005602239,
"recall": 0.9285714285714286,
"f1": 0.9226432439812495,
"num_samples": 574.0
}
# total samples 574
# samples for OSS models: 544 (94.8%)
Performance on samples predicted for codeLlama-34b: 3.87
Performance on samples predicted for gpt-4: 3.55
Note: For our dataset, a small logistic regression model is good enough to perform the routing. But if your use case is more complex, consider training a more complex model, like a BERT-based classifier to perform the classification. These models are still small enough that wouldn’t introduce too much latency. Be sure to check out this guide if you want to learn how to train and deploy supervised deep learning models.
LinkServing
Now we're ready to start serving our Ray Assistant using our best configuration. We're going to use Ray Serve with FastAPI to develop and scale our service. First, we'll define some data structures like Query and Answer to represent the inputs and outputs to our service. We will also define a small function to load our index (assumes that the respective SQL dump file already exists). Finally, we can define our QueryAgent and use it to serve POST requests with the query. And we can serve our agent at any deployment scale we wish using the @serve.deployment decorator where we can specify the number of replicas, compute resources, etc.
1# Initialize application
2app = FastAPI()
3
4@serve.deployment(route_prefix="/", num_replicas=1, ray_actor_options={"num_cpus": 6, "num_gpus": 1})
5@serve.ingress(app)
6class RayAssistantDeployment:
7 def __init__(self, chunk_size, chunk_overlap, num_chunks,
8 embedding_model_name, embedding_dim,
9 use_lexical_search, lexical_search_k,
10 use_reranking, rerank_threshold, rerank_k,
11 llm, sql_dump_fp=None):
12
13 # Set up
14 chunks = build_or_load_index(
15 embedding_model_name=embedding_model_name,
16 embedding_dim=embedding_dim,
17 chunk_size=chunk_size,
18 chunk_overlap=chunk_overlap,
19 sql_dump_fp=sql_dump_fp,
20 )
21
22 # Lexical index
23 lexical_index = None
24 self.lexical_search_k = lexical_search_k
25 if use_lexical_search:
26 texts = [re.sub(r"[^a-zA-Z0-9]", " ", chunk[1]).lower().split() for chunk in chunks]
27 lexical_index = BM25Okapi(texts)
28
29 # Reranker
30 reranker = None
31 self.rerank_threshold = rerank_threshold
32 self.rerank_k = rerank_k
33 if use_reranking:
34 reranker_fp = Path(EFS_DIR, "reranker.pkl")
35 with open(reranker_fp, "rb") as file:
36 reranker = pickle.load(file)
37
38 # Query agent
39 self.num_chunks = num_chunks
40 system_content = "Answer the query using the context provided. Be succinct. " \
41 "Contexts are organized in a list of dictionaries [{'text': <context>}, {'text': <context>}, ...]. " \
42 "Feel free to ignore any contexts in the list that don't seem relevant to the query. "
43 self.oss_agent = QueryAgent(
44 embedding_model_name=embedding_model_name,
45 chunks=chunks,
46 lexical_index=lexical_index,
47 reranker=reranker,
48 llm=llm,
49 max_context_length=MAX_CONTEXT_LENGTHS[llm],
50 system_content=system_content)
51 self.gpt_agent = QueryAgent(
52 embedding_model_name=embedding_model_name,
53 chunks=chunks,
54 lexical_index=lexical_index,
55 reranker=reranker,
56 llm="gpt-4",
57 max_context_length=MAX_CONTEXT_LENGTHS["gpt-4"],
58 system_content=system_content)
59
60 # Router
61 router_fp = Path(EFS_DIR, "router.pkl")
62 with open(router_fp, "rb") as file:
63 self.router = pickle.load(file)
64
65 @app.post("/query")
66 def query(self, query: Query) -> Answer:
67 use_oss_agent = self.router.predict([query.query])[0]
68 agent = self.oss_agent if use_oss_agent else self.gpt_agent
69 result = agent(
70 query=query.query, num_chunks=self.num_chunks,
71 lexical_search_k=self.lexical_search_k,
72 rerank_threshold=self.rerank_threshold,
73 rerank_k=self.rerank_k,
74 stream=False)
75 return Answer.parse_obj(result)1# Deploy the Ray Serve application.
2deployment = RayAssistantDeployment.bind(
3 chunk_size=700,
4 chunk_overlap=50,
5 num_chunks=9,
6 embedding_model_name=os.environ["RAY_ASSISTANT_EMBEDDING_MODEL"],
7 embedding_dim=EMBEDDING_DIMENSIONS["thenlper/gte-large"],
8 use_lexical_search=False,
9 lexical_search_k=0,
10 use_reranking=True,
11 rerank_threshold=0.9,
12 rerank_k=9,
13 llm="mistralai/Mixtral-8x7B-Instruct-v0.1",
14 sql_dump_fp=Path(os.environ["RAY_ASSISTANT_INDEX"]))
15serve.run(deployment)And with our application served, we’re ready to query it!
1# Inference
2data = {"query": "What is the default batch size for map_batches?"}
3response = requests.post("http://127.0.0.1:8000/query", json=data)
4print(response.text)
5{
'question': 'What is the default batch size for map_batches?',
'sources': [
'ray.data.Dataset.map_batches — Ray 2.7.1',
'Transforming Data — Ray 2.7.1',
...
],
'answer': 'The default batch size for map_batches is 4096.',
'llm': 'mistralai/Mixtral-8x7B-Instruct-v0.1'
}
Note: As we can see, Ray Serve makes model composition extremely easy and we could continue to make this even more fine-grained with more workflow logic.
Once our application is served, we’re free to use it anywhere we want. For example, we use it as a bot on our Slack channels and as a widget on our docs page (public release coming soon). We can use this to collect feedback from our users to continually improve the application (fine-tuning, UI/UX, etc.).
LinkData flywheel
Creating an application like this is not a one-time task. It's extremely important that we continue to iterate and keep our application up to date. This includes continually reindexing our data so that our application is working with the most up-to-date information. As well as rerunning our experiments to see if any of the decisions need to be altered. This process of continuous iteration can be achieved by mapping our workflows to CI/CD pipelines.
A key part of iteration that goes beyond automated reindexing, evaluation, etc. involves fixing our data itself. In fact, we found that this is the most impactful lever (way beyond our retrieval and generation optimizations above) we could control. Here is an example workflow we've settled on:
Users use the RAG application to ask questions about the product.
Use feedback (👍/👎, visited source pages, top-k cosine scores, etc.) to identify underperforming queries.
Inspect the retrieved resources, tokenization, etc. to decide if it's a shortcoming of retrieval, generation or the underlying data source.
If something in the data can be improved, separated into sections/pages, etc. → fix it!
Evaluate (and add to test suite) on previously underperforming queries.
Reindex and deploy a new, potentially further optimized, version of the application.
LinkImpact
LinkProducts and productivity
Building an LLM application like this has had a tremendous impact on our products and company. There were expected 1st order impacts in overall developer and user adoption for our products. The capability to interact and solve problems that our users experience in a self-serve and immediate manner is the type of feature that would improve the experience of any product. It makes it significantly easier for people to succeed and it elevated the perception around LLM applications from a nice-to-have to a must-have.
LinkFoundational agents
However, there were also some 2nd order impacts that we didn’t immediately realize. For example, when we further inspected user queries that yielded poor scores, often the issue existed because of a gap in our documentation. When we made the fix (ex. added the appropriate section to our docs), this improved our product and the LLM application itself — creating a very valuable feedback flywheel. Furthermore, when internal teams learned of the capabilities of our LLM application, this generated the development of highly valuable LLM applications that depend on this Ray docs LLM application as one of its foundational agents that it uses to perform its tasks.
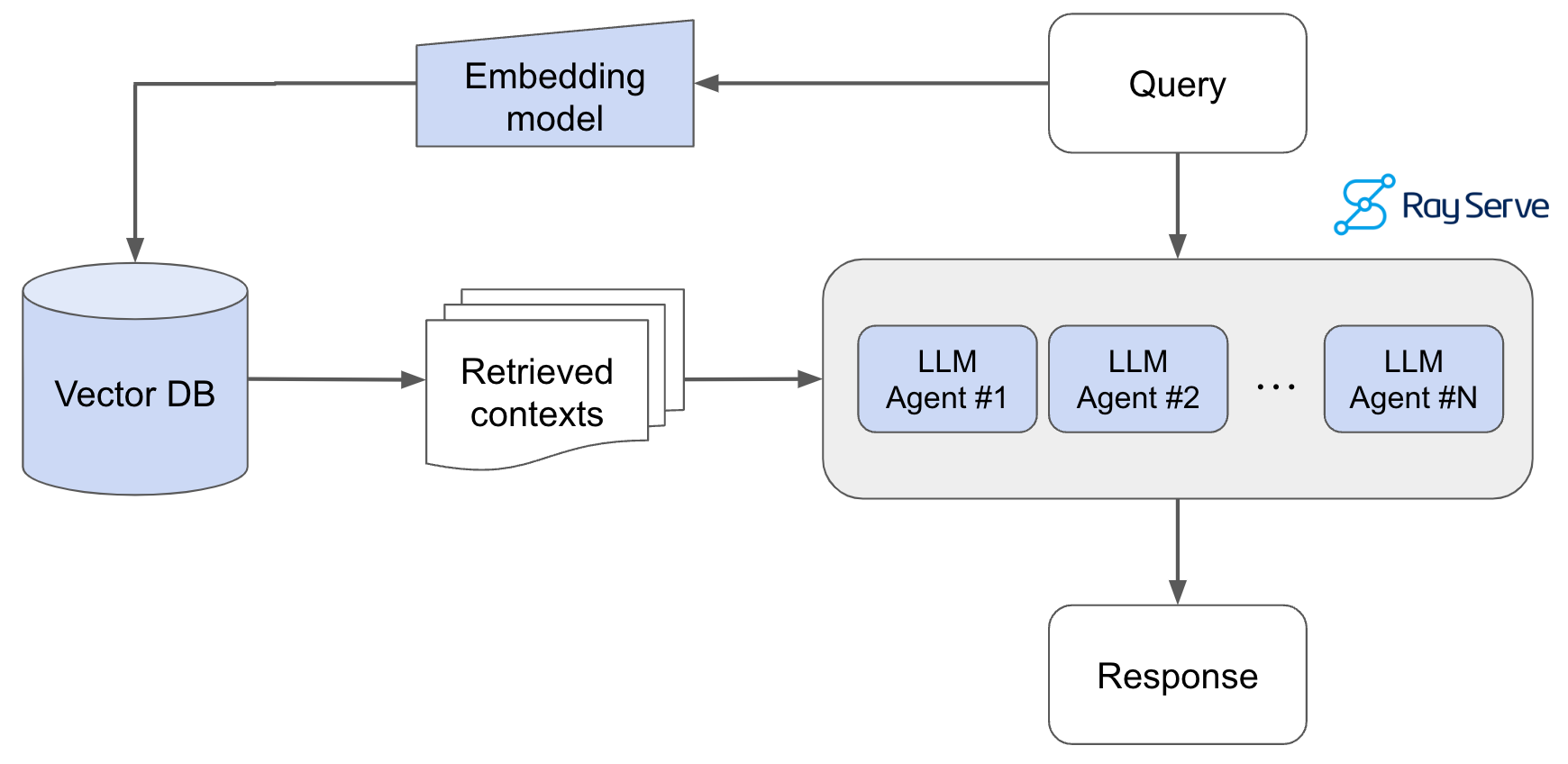
For example, we’ve internally developed a feature called Anyscale Doctor that helps developers diagnose and debug issues during development. Issues in code can be caused by a variety of reasons but when the issue is Ray related, the LLM application we built here is called to aid in resolving the particular issue.
LinkLearn more
If your team is investing heavily in developing LLM applications, reach out to us to learn more about how Ray and Anyscale can help you scale and productionize everything.
Start serving (+fine-tuning) OSS LLMs with Anyscale Endpoints ($1/M tokens for Llama-2-70b) w/ 1M free tokens trial.
If you need to deploy on your own private cloud, check out Anyscale Private Endpoints.
Learn more about how companies like OpenAI, Netflix, Pinterest, Verizon, Instacart and others leverage Ray and Anyscale for their AI workloads at the Ray Summit.


